Mass-spectrometry-based spatial proteomics data analysis using pRoloc and pRolocdata
- PMID: 24413670
- PMCID: PMC3998135
- DOI: 10.1093/bioinformatics/btu013
Mass-spectrometry-based spatial proteomics data analysis using pRoloc and pRolocdata
Abstract
Motivation: Experimental spatial proteomics, i.e. the high-throughput assignment of proteins to sub-cellular compartments based on quantitative proteomics data, promises to shed new light on many biological processes given adequate computational tools.
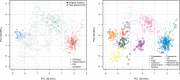
Results: Here we present pRoloc, a complete infrastructure to support and guide the sound analysis of quantitative mass-spectrometry-based spatial proteomics data. It provides functionality for unsupervised and supervised machine learning for data exploration and protein classification and novelty detection to identify new putative sub-cellular clusters. The software builds upon existing infrastructure for data management and data processing.
Figures
Similar articles
-
A Bioconductor workflow for the Bayesian analysis of spatial proteomics.F1000Res. 2019 Apr 11;8:446. doi: 10.12688/f1000research.18636.1. eCollection 2019. F1000Res. 2019. PMID: 31119032 Free PMC article.
-
A Bioconductor workflow for processing and analysing spatial proteomics data.F1000Res. 2016 Dec 28;5:2926. doi: 10.12688/f1000research.10411.2. eCollection 2016. F1000Res. 2016. PMID: 30079225 Free PMC article.
-
The effect of organelle discovery upon sub-cellular protein localisation.J Proteomics. 2013 Aug 2;88:129-40. doi: 10.1016/j.jprot.2013.02.019. Epub 2013 Mar 21. J Proteomics. 2013. PMID: 23523639
-
Methods and Algorithms for Quantitative Proteomics by Mass Spectrometry.Methods Mol Biol. 2020;2051:161-197. doi: 10.1007/978-1-4939-9744-2_7. Methods Mol Biol. 2020. PMID: 31552629 Review.
-
Application of Machine Learning in Spatial Proteomics.J Chem Inf Model. 2022 Dec 12;62(23):5875-5895. doi: 10.1021/acs.jcim.2c01161. Epub 2022 Nov 15. J Chem Inf Model. 2022. PMID: 36378082 Review.
Cited by
-
Operation of a TCA cycle subnetwork in the mammalian nucleus.Sci Adv. 2022 Sep 2;8(35):eabq5206. doi: 10.1126/sciadv.abq5206. Epub 2022 Aug 31. Sci Adv. 2022. PMID: 36044572 Free PMC article.
-
Spatial proteomics reveals subcellular reorganization in human keratinocytes exposed to UVA light.iScience. 2022 Mar 16;25(4):104093. doi: 10.1016/j.isci.2022.104093. eCollection 2022 Apr 15. iScience. 2022. PMID: 35372811 Free PMC article.
-
Assessing sub-cellular resolution in spatial proteomics experiments.Curr Opin Chem Biol. 2019 Feb;48:123-149. doi: 10.1016/j.cbpa.2018.11.015. Epub 2018 Dec 14. Curr Opin Chem Biol. 2019. PMID: 30711721 Free PMC article. Review.
-
Semi-Supervised Non-Parametric Bayesian Modelling of Spatial Proteomics.Ann Appl Stat. 2022 Dec 1;16(4):22-aoas1603. doi: 10.1214/22-AOAS1603. eCollection 2022 Dec 1. Ann Appl Stat. 2022. PMID: 36507469 Free PMC article.
-
Comparative Analysis of T-Cell Spatial Proteomics and the Influence of HIV Expression.Mol Cell Proteomics. 2022 Mar;21(3):100194. doi: 10.1016/j.mcpro.2022.100194. Epub 2022 Jan 8. Mol Cell Proteomics. 2022. PMID: 35017099 Free PMC article.
References
-
- Breckels L, et al. The effect of organelle discovery upon sub-cellular protein localisation. J. Proteom. 2013;88:129–140. - PubMed
-
- Courty N, et al. Perturbo: a new classification algorithm based on the spectrum perturbations of the laplace-beltrami operator. In: Gunopulos D, et al., editors. The Proceedings of ECML/PKDD (1) 2011. Vol. 6911 of Lecture Notes in Computer Science, pp. 359–374. Springer-Verlag, Berlin Heidelberg.
-
- Gatto L, Christoforou A. Using R and Bioconductor for proteomics data analysis. Biochim. Biophys. Acta. 2013;1844(1 Pt A):42–51. - PubMed
-
- Gatto L, Lilley KS. MSnbase – an R/Bioconductor package for isobaric tagged mass spectrometry data visualization, processing and quantitation. Bioinformatics. 2012;28:288–289. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

