Incorporating DNA shearing in standard affinity purification allows simultaneous identification of both soluble and chromatin-bound interaction partners
- PMID: 24412199
- PMCID: PMC4128382
- DOI: 10.1016/j.jprot.2013.12.022
Incorporating DNA shearing in standard affinity purification allows simultaneous identification of both soluble and chromatin-bound interaction partners
Abstract
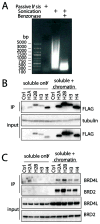
Affinity purification coupled to mass spectrometry (AP-MS) is an effective means of identifying protein-protein interactions to better understand biological functions. However, issues associated with sample preparation still limit the success of AP-MS for specific classes of proteins, including those associated with chromatin that exhibit overall poor solubility in the protocols normally used for AP-MS analysis. Here, we wanted to provide a generally applicable method to simultaneously identify interactors for the chromatin-bound and the soluble fractions of a given bait protein. Using four FLAG-tagged canonical histone proteins (H2A, H2B, H3.1 and H4) we demonstrate that the chromatin solubility issue can be robustly alleviated by fragmenting DNA prior to AP-MS using a combination of sonication and nuclease treatment. We show that - in comparison to a commonly used AP-MS method - our optimized protocol greatly improves the recovery of chromatin-associated interactors for core histones. Critically, this is achieved while preserving the interaction partners associated with the soluble portion of the histones. Detailed protocols amenable to the study of both histone and non-histone baits are presented here.
Biological significance: This manuscript describes workflow improvements to enable the recovery of chromatin-bound interactors by affinity purification coupled to mass spectrometry (AP-MS). This is significant, as most of the high-throughput studies to date can only monitor protein-protein interactions for soluble (not bound to chromatin) components. By consequence, we still poorly understand how protein complexes form on chromatin, which greatly hampers our understanding of gene expression. Using core histones as test cases, we show here a simple and universally applicable workflow that permits the identification of chromatin-bound protein-protein interactions. As exemplified in our manuscript, this revised protocol should result in a much deeper understanding of chromatin biology. This article is part of a Special Issue: Can Proteomics Fill the Gap Between Genomics and Phenotypes?
Keywords: Affinity purification coupled to mass spectrometry; Chromatin; Protein–protein interactions; Systems biology.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes.J Proteomics. 2015 Apr 6;118:81-94. doi: 10.1016/j.jprot.2014.09.011. Epub 2014 Oct 2. J Proteomics. 2015. PMID: 25281560 Free PMC article.
-
Parallel Exploration of Interaction Space by BioID and Affinity Purification Coupled to Mass Spectrometry.Methods Mol Biol. 2017;1550:115-136. doi: 10.1007/978-1-4939-6747-6_10. Methods Mol Biol. 2017. PMID: 28188527
-
Combined proximity labeling and affinity purification-mass spectrometry workflow for mapping and visualizing protein interaction networks.Nat Protoc. 2020 Oct;15(10):3182-3211. doi: 10.1038/s41596-020-0365-x. Epub 2020 Aug 10. Nat Protoc. 2020. PMID: 32778839
-
Advanced methods for the analysis of chromatin-associated proteins.Physiol Genomics. 2014 Jul 1;46(13):441-7. doi: 10.1152/physiolgenomics.00041.2014. Epub 2014 May 6. Physiol Genomics. 2014. PMID: 24803678 Free PMC article. Review.
-
Computational and informatics strategies for identification of specific protein interaction partners in affinity purification mass spectrometry experiments.Proteomics. 2012 May;12(10):1639-55. doi: 10.1002/pmic.201100537. Proteomics. 2012. PMID: 22611043 Free PMC article. Review.
Cited by
-
A Nutrient-Based Cellular Model to Characterize Acetylation-Dependent Protein-Protein Interactions.Front Mol Biosci. 2022 Mar 23;9:831758. doi: 10.3389/fmolb.2022.831758. eCollection 2022. Front Mol Biosci. 2022. PMID: 35402505 Free PMC article.
-
High-Efficiency Enrichment by Saturating Nanoliters of Protein Affinity Media.Anal Chem. 2023 Oct 31;95(43):15884-15892. doi: 10.1021/acs.analchem.3c01736. Epub 2023 Oct 18. Anal Chem. 2023. PMID: 37851921 Free PMC article.
-
Assessing histone demethylase inhibitors in cells: lessons learned.Epigenetics Chromatin. 2017 Mar 1;10:9. doi: 10.1186/s13072-017-0116-6. eCollection 2017. Epigenetics Chromatin. 2017. PMID: 28265301 Free PMC article.
-
Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes.J Proteomics. 2015 Apr 6;118:81-94. doi: 10.1016/j.jprot.2014.09.011. Epub 2014 Oct 2. J Proteomics. 2015. PMID: 25281560 Free PMC article.
-
Meet the neighbors: Mapping local protein interactomes by proximity-dependent labeling with BioID.Proteomics. 2016 Oct;16(19):2503-2518. doi: 10.1002/pmic.201600123. Epub 2016 Jul 27. Proteomics. 2016. PMID: 27329485 Free PMC article. Review.
References
-
- Lambert JP, Pawson T, Gingras AC. Mapping physical interactions within chromatin by proteomic approaches. Proteomics. 2012;12:1609–22. - PubMed
-
- Sidoli S, Cheng L, Jensen ON. Proteomics in chromatin biology and epigenetics: Elucidation of post-translational modifications of histone proteins by mass spectrometry. J Proteomics. 2012;75:3419–33. - PubMed
-
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

