RNA-Seq derived identification of differential transcription in the chrysanthemum leaf following inoculation with Alternaria tenuissima
- PMID: 24387266
- PMCID: PMC3890596
- DOI: 10.1186/1471-2164-15-9
RNA-Seq derived identification of differential transcription in the chrysanthemum leaf following inoculation with Alternaria tenuissima
Abstract
Background: A major production constraint on the important ornamental species chrysanthemum is black spot which is caused by the necrotrophic fungus Alternaria tenuissima. The molecular basis of host resistance to A. tenuissima has not been studied as yet in any detail. Here, high throughput sequencing was taken to characterize the transcriptomic response of the chrysanthemum leaf to A. tenuissima inoculation.
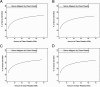
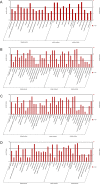
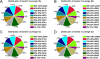
Results: The transcriptomic data was acquired using RNA-Seq technology, based on the Illumina HiSeq™ 2000 platform. Four different libraries derived from two sets of leaves harvested from either inoculated or mock-inoculated plants were characterized. Over seven million clean reads were generated from each library, each corresponding to a coverage of >350,000 nt. About 70% of the reads could be mapped to a set of chrysanthemum unigenes. Read frequency was used as a measure of transcript abundance and therefore as an identifier of differential transcription in the four libraries. The differentially transcribed genes identified were involved in photosynthesis, pathogen recognition, reactive oxygen species generation, cell wall modification and phytohormone signalling; in addition, a number of varied transcription factors were identified. A selection of 23 of the genes was transcription-profiled using quantitative RT-PCR to validate the RNA-Seq output.
Conclusions: A substantial body of chrysanthemum transcriptomic sequence was generated, which led to a number of insights into the molecular basis of the host response to A. tenuissima infection. Although most of the differentially transcribed genes were up-regulated by the presence of the pathogen, those involved in photosynthesis were down-regulated.
Figures
Similar articles
-
A transcriptomic analysis of Chrysanthemum nankingense provides insights into the basis of low temperature tolerance.BMC Genomics. 2014 Oct 3;15(1):844. doi: 10.1186/1471-2164-15-844. BMC Genomics. 2014. PMID: 25277256 Free PMC article.
-
CmWRKY15 Facilitates Alternaria tenuissima Infection of Chrysanthemum.PLoS One. 2015 Nov 24;10(11):e0143349. doi: 10.1371/journal.pone.0143349. eCollection 2015. PLoS One. 2015. PMID: 26600125 Free PMC article.
-
Interaction of roses with a biotrophic and a hemibiotrophic leaf pathogen leads to differences in defense transcriptome activation.Plant Mol Biol. 2019 Mar;99(4-5):299-316. doi: 10.1007/s11103-018-00818-2. Epub 2019 Jan 31. Plant Mol Biol. 2019. PMID: 30706286
-
Transcriptomic analysis of differentially expressed genes in the floral transition of the summer flowering chrysanthemum.BMC Genomics. 2016 Aug 24;17(1):673. doi: 10.1186/s12864-016-3024-4. BMC Genomics. 2016. PMID: 27552984 Free PMC article.
-
Dual species dynamic transcripts reveal the interaction mechanisms between Chrysanthemum morifolium and Alternaria alternata.BMC Genomics. 2021 Jul 9;22(1):523. doi: 10.1186/s12864-021-07709-9. BMC Genomics. 2021. PMID: 34243707 Free PMC article.
Cited by
-
Comparative Transcriptome Analysis of Isoetes Sinensis Under Terrestrial and Submerged Conditions.Plant Mol Biol Report. 2016;34:136-145. doi: 10.1007/s11105-015-0906-6. Epub 2015 Jun 27. Plant Mol Biol Report. 2016. PMID: 26843780 Free PMC article.
-
Transcriptomic analysis unveils alterations in the genetic expression profile of tree peony (Paeonia suffruticosa Andrews) infected by Alternaria alternata.BMC Genomics. 2024 Sep 14;25(1):861. doi: 10.1186/s12864-024-10784-3. BMC Genomics. 2024. PMID: 39277723 Free PMC article.
-
RNA-seq Transcriptome Response of Flax (Linum usitatissimum L.) to the Pathogenic Fungus Fusarium oxysporum f. sp. lini.Front Plant Sci. 2016 Nov 24;7:1766. doi: 10.3389/fpls.2016.01766. eCollection 2016. Front Plant Sci. 2016. PMID: 27933082 Free PMC article.
-
De novo transcriptome assembly of pummelo and molecular marker development.PLoS One. 2015 Mar 23;10(3):e0120615. doi: 10.1371/journal.pone.0120615. eCollection 2015. PLoS One. 2015. PMID: 25799271 Free PMC article.
-
A temporal gene expression map of Chrysanthemum leaves infected with Alternaria alternata reveals different stages of defense mechanisms.Hortic Res. 2020 Mar 1;7:23. doi: 10.1038/s41438-020-0245-0. eCollection 2020. Hortic Res. 2020. PMID: 32140232 Free PMC article.
References
-
- Shibata M. Importance of genetic transformation in ornamental plant breeding. Plant Biotechnol. 2008;15(1):3–8. doi: 10.5511/plantbiotechnology.25.3. - DOI
-
- Teixeira da Silva JA, Shinoyama H, Aida R, Matsushita Y, Raj SK, Chen F. Chrysanthemum Biotechnology: Quo vadis? Crit Rev Plant Sci. 2013;15(1):21–52. doi: 10.1080/07352689.2012.696461. - DOI
-
- Xu G, Liu Y, Chen S, Chen F. Potential structural and biochemical mechanisms of compositae wild species resistance to Alternaria tenuissima. Russ J Plant Physl+ 2011;15(3):491–497. doi: 10.1134/S1021443711030216. - DOI
-
- Deng Y, Chen S, Chang Q, Wang H, Chen F. The chrysanthemum × Artemisia vulgaris intergeneric hybrid has better rooting ability and higher resistance to alternaria leaf spot than its chrysanthemum parent. Sci Hortic-Amsterdam. 2012;15:185–190.
-
- Mirkova E, Konstantinova P. First report of Alternaria leaf spot on gerbera (Gerbera jamesonii H. Bolux ex JD Hook) in Bulgaria. J Phytopathol. 2003;15(6):323–328.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

