Overcoming antigenic diversity by enhancing the immunogenicity of conserved epitopes on the malaria vaccine candidate apical membrane antigen-1
- PMID: 24385910
- PMCID: PMC3873463
- DOI: 10.1371/journal.ppat.1003840
Overcoming antigenic diversity by enhancing the immunogenicity of conserved epitopes on the malaria vaccine candidate apical membrane antigen-1
Erratum in
- PLoS Pathog. 2014 Jan;10(1). doi:10.1371/annotation/be0d67d1-314f-4654-8a2f-b668d1e32bbd. Ge, Xiopeng [corrected to Ge, Xiaopeng]
Abstract
Malaria vaccine candidate Apical Membrane Antigen-1 (AMA1) induces protection, but only against parasite strains that are closely related to the vaccine. Overcoming the AMA1 diversity problem will require an understanding of the structural basis of cross-strain invasion inhibition. A vaccine containing four diverse allelic proteins 3D7, FVO, HB3 and W2mef (AMA1 Quadvax or QV) elicited polyclonal rabbit antibodies that similarly inhibited the invasion of four vaccine and 22 non-vaccine strains of P. falciparum. Comparing polyclonal anti-QV with antibodies against a strain-specific, monovalent, 3D7 AMA1 vaccine revealed that QV induced higher levels of broadly inhibitory antibodies which were associated with increased conserved face and domain-3 responses and reduced domain-2 response. Inhibitory monoclonal antibodies (mAb) raised against the QV reacted with a novel cross-reactive epitope at the rim of the hydrophobic trough on domain-1; this epitope mapped to the conserved face of AMA1 and it encompassed the 1e-loop. MAbs binding to the 1e-loop region (1B10, 4E8 and 4E11) were ∼10-fold more potent than previously characterized AMA1-inhibitory mAbs and a mode of action of these 1e-loop mAbs was the inhibition of AMA1 binding to its ligand RON2. Unlike the epitope of a previously characterized 3D7-specific mAb, 1F9, the 1e-loop inhibitory epitope was partially conserved across strains. Another novel mAb, 1E10, which bound to domain-3, was broadly inhibitory and it blocked the proteolytic processing of AMA1. By itself mAb 1E10 was weakly inhibitory but it synergized with a previously characterized, strain-transcending mAb, 4G2, which binds close to the hydrophobic trough on the conserved face and inhibits RON2 binding to AMA1. Novel inhibition susceptible regions and epitopes, identified here, can form the basis for improving the antigenic breadth and inhibitory response of AMA1 vaccines. Vaccination with a few diverse antigenic proteins could provide universal coverage by redirecting the immune response towards conserved epitopes.
Conflict of interest statement
SD has been named on an AMA1 related US patent. This does not alter our adherence to all PLOS policies on sharing data and materials.
Figures

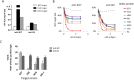


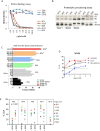
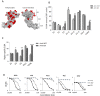
Similar articles
-
Defining the antigenic diversity of Plasmodium falciparum apical membrane antigen 1 and the requirements for a multi-allele vaccine against malaria.PLoS One. 2012;7(12):e51023. doi: 10.1371/journal.pone.0051023. Epub 2012 Dec 5. PLoS One. 2012. PMID: 23227229 Free PMC article.
-
Structural basis of antigenic escape of a malaria vaccine candidate.Proc Natl Acad Sci U S A. 2007 Jul 24;104(30):12488-93. doi: 10.1073/pnas.0701464104. Epub 2007 Jul 16. Proc Natl Acad Sci U S A. 2007. PMID: 17636123 Free PMC article.
-
Rapid and precise epitope mapping of monoclonal antibodies against Plasmodium falciparum AMA1 by combined phage display of fragments and random peptides.Protein Eng. 2001 Sep;14(9):691-8. doi: 10.1093/protein/14.9.691. Protein Eng. 2001. PMID: 11707616
-
Apical membrane antigen 1: a malaria vaccine candidate in review.Trends Parasitol. 2008 Feb;24(2):74-84. doi: 10.1016/j.pt.2007.12.002. Epub 2008 Jan 15. Trends Parasitol. 2008. PMID: 18226584 Review.
-
Apical membrane antigen 1 as an anti-malarial drug target.Curr Top Med Chem. 2011;11(16):2039-47. doi: 10.2174/156802611796575885. Curr Top Med Chem. 2011. PMID: 21619512 Review.
Cited by
-
Analysis of the dose-dependent stage-specific in vitro efficacy of a multi-stage malaria vaccine candidate cocktail.Malar J. 2016 May 17;15(1):279. doi: 10.1186/s12936-016-1328-0. Malar J. 2016. PMID: 27188716 Free PMC article.
-
Low levels of polymorphisms and no evidence for diversifying selection on the Plasmodium knowlesi Apical Membrane Antigen 1 gene.PLoS One. 2015 Apr 16;10(4):e0124400. doi: 10.1371/journal.pone.0124400. eCollection 2015. PLoS One. 2015. PMID: 25881166 Free PMC article.
-
Structure-based design of a strain transcending AMA1-RON2L malaria vaccine.Nat Commun. 2023 Sep 2;14(1):5345. doi: 10.1038/s41467-023-40878-7. Nat Commun. 2023. PMID: 37660103 Free PMC article.
-
Use of immunodampening to overcome diversity in the malarial vaccine candidate apical membrane antigen 1.Infect Immun. 2014 Nov;82(11):4707-17. doi: 10.1128/IAI.02061-14. Epub 2014 Aug 25. Infect Immun. 2014. PMID: 25156737 Free PMC article.
-
Variations in the quality of malaria-specific antibodies with transmission intensity in a seasonal malaria transmission area of Northern Ghana.PLoS One. 2017 Sep 25;12(9):e0185303. doi: 10.1371/journal.pone.0185303. eCollection 2017. PLoS One. 2017. PMID: 28945794 Free PMC article.
References
-
- Sachs J, Malaney P (2002) The economic and social burden of malaria. Nature 415: 680–685. - PubMed
-
- Tham WH, Healer J, Cowman AF (2012) Erythrocyte and reticulocyte binding-like proteins of Plasmodium falciparum. Trends Parasitol 28: 23–30. - PubMed
-
- Remarque EJ, Faber BW, Kocken CH, Thomas AW (2008) Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol 24: 74–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

