Genetic control of specificity to steroid-triggered responses in Drosophila
- PMID: 24374353
- PMCID: PMC3948805
- DOI: 10.1534/genetics.113.159707
Genetic control of specificity to steroid-triggered responses in Drosophila
Abstract
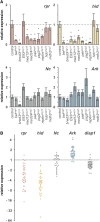
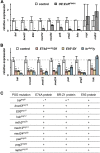
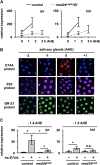
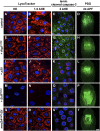
Steroid hormones trigger a wide variety of biological responses through stage- and tissue-specific activation of target gene expression. The mechanisms that provide specificity to systemically released pulses of steroids, however, remain poorly understood. We previously completed a forward genetic screen for mutations that disrupt the destruction of larval salivary glands during metamorphosis in Drosophila melanogaster, a process triggered by the steroid hormone 20-hydroxyecdysone (ecdysone). Here, we characterize 10 complementation groups mapped to genes from this screen. Most of these mutations disrupt the ecdysone-induced expression of death activators, thereby failing to initiate tissue destruction. However, other responses to ecdysone, even within salivary glands, occur normally in mutant animals. Many of these newly identified regulators of ecdysone signaling, including brwd3, med12, med24, pak, and psg2, represent novel components of the ecdysone-triggered transcriptional hierarchy. These genes function combinatorially to provide specificity to ecdysone pulses, amplifying the hormonal cue in a stage-, tissue-, and target gene-specific manner. Most of the ecdysone response genes identified in this screen encode homologs of mammalian nuclear receptor coregulators, demonstrating an unexpected degree of functional conservation in the mechanisms that regulate steroid signaling between insects and mammals.
Keywords: cell death; ecdysone; salivary glands; specificity; transcription.
Figures


Similar articles
-
Fhos encodes a Drosophila Formin-like protein participating in autophagic programmed cell death.Genesis. 2012 Sep;50(9):672-84. doi: 10.1002/dvg.22025. Epub 2012 Apr 25. Genesis. 2012. PMID: 22422652
-
Control of target gene specificity during metamorphosis by the steroid response gene E93.Proc Natl Acad Sci U S A. 2012 Feb 21;109(8):2949-54. doi: 10.1073/pnas.1117559109. Epub 2012 Feb 2. Proc Natl Acad Sci U S A. 2012. PMID: 22308414 Free PMC article.
-
Drosophila E93 promotes adult development and suppresses larval responses to ecdysone during metamorphosis.Dev Biol. 2022 Jan;481:104-115. doi: 10.1016/j.ydbio.2021.10.001. Epub 2021 Oct 11. Dev Biol. 2022. PMID: 34648816 Free PMC article.
-
Ecdysone signaling cascade and regulation of Drosophila metamorphosis.Arch Insect Biochem Physiol. 1996;33(3-4):231-44. doi: 10.1002/(SICI)1520-6327(1996)33:3/4<231::AID-ARCH5>3.0.CO;2-V. Arch Insect Biochem Physiol. 1996. PMID: 8913033 Review.
-
What goes up must come down: transcription factors have their say in making ecdysone pulses.Curr Top Dev Biol. 2013;103:35-71. doi: 10.1016/B978-0-12-385979-2.00002-2. Curr Top Dev Biol. 2013. PMID: 23347515 Review.
Cited by
-
Systemic and local effect of the Drosophila headcase gene and its role in stress protection of Adult Progenitor Cells.PLoS Genet. 2021 Feb 8;17(2):e1009362. doi: 10.1371/journal.pgen.1009362. eCollection 2021 Feb. PLoS Genet. 2021. PMID: 33556132 Free PMC article.
-
Next-Generation Sequencing Reveals Increased Anti-oxidant Response and Ecdysone Signaling in STAT Supercompetitors in Drosophila.G3 (Bethesda). 2019 Aug 8;9(8):2609-2622. doi: 10.1534/g3.119.400345. G3 (Bethesda). 2019. PMID: 31227525 Free PMC article.
-
Diverse Hormone Response Networks in 41 Independent Drosophila Cell Lines.G3 (Bethesda). 2016 Jan 15;6(3):683-94. doi: 10.1534/g3.115.023366. G3 (Bethesda). 2016. PMID: 26772746 Free PMC article.
-
Eaten to death.FEBS J. 2014 Dec;281(24):5411-7. doi: 10.1111/febs.13114. Epub 2014 Nov 10. FEBS J. 2014. PMID: 25323556 Free PMC article. Review.
-
A cryptic Tudor domain links BRWD2/PHIP to COMPASS-mediated histone H3K4 methylation.Genes Dev. 2017 Oct 1;31(19):2003-2014. doi: 10.1101/gad.305201.117. Genes Dev. 2017. PMID: 29089422 Free PMC article.
References
-
- Andres A. J., Thummel C. S., 1994. Methods for quantitative analysis of transcription in larvae and prepupae. Methods Cell Biol. 44: 565–573. - PubMed
-
- Ashburner M., 1974. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. II. The effects of inhibitors of protein synthesis. Dev. Biol. 39: 141–157. - PubMed
-
- Ashburner M., 1989. Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Ashburner M., Chihara C., Meltzer P., Richards G., 1974. Temporal control of puffing activity in polytene chromosomes. Cold Spring Harb. Symp. Quant. Biol. 38: 655–662. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

