SUMOylation of the polycomb group protein L3MBTL2 facilitates repression of its target genes
- PMID: 24369422
- PMCID: PMC3950706
- DOI: 10.1093/nar/gkt1317
SUMOylation of the polycomb group protein L3MBTL2 facilitates repression of its target genes
Abstract
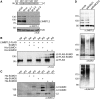
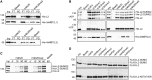
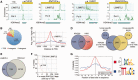
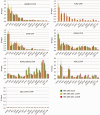
Lethal(3) malignant brain tumour like 2 (L3MBTL2) is an integral component of the polycomb repressive complex 1.6 (PRC1.6) and has been implicated in transcriptional repression and chromatin compaction. Here, we show that L3MBTL2 is modified by SUMO2/3 at lysine residues 675 and 700 close to the C-terminus. SUMOylation of L3MBTL2 neither affected its repressive activity in reporter gene assays nor it's binding to histone tails in vitro. In order to analyse whether SUMOylation affects binding of L3MBTL2 to chromatin, we performed ChIP-Seq analysis with chromatin of wild-type HEK293 cells and with chromatin of HEK293 cells stably expressing either FLAG-tagged SUMOylation-competent or SUMOylation-defective L3MBTL2. Wild-type FLAG-L3MBTL2 and the SUMOylation-defective FLAG-L3MBTL2 K675/700R mutant essentially occupied the same sites as endogenous L3MBTL2 suggesting that SUMOylation of L3MBTL2 does not affect chromatin binding. However, a subset of L3MBTL2-target genes, particularly those with low L3MBTL2 occupancy including pro-inflammatory genes, was de-repressed in cells expressing the FLAG-L3MBTL2 K675/700R mutant. Finally, we provide evidence that SUMOylation of L3MBTL2 facilitates repression of these PRC1.6-target genes by balancing the local H2Aub1 levels established by the ubiquitinating enzyme RING2 and the de-ubiquitinating PR-DUB complex.
Figures



Similar articles
-
MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6.PLoS Genet. 2018 Jan 30;14(1):e1007193. doi: 10.1371/journal.pgen.1007193. eCollection 2018 Jan. PLoS Genet. 2018. PMID: 29381691 Free PMC article.
-
L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure.Mol Cell. 2011 May 20;42(4):438-50. doi: 10.1016/j.molcel.2011.04.004. Mol Cell. 2011. PMID: 21596310 Free PMC article.
-
The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development.Cell Stem Cell. 2012 Sep 7;11(3):319-32. doi: 10.1016/j.stem.2012.06.002. Epub 2012 Jul 5. Cell Stem Cell. 2012. PMID: 22770845 Free PMC article.
-
L3MBTL2 regulates chromatin remodeling during spermatogenesis.Cell Death Differ. 2019 Nov;26(11):2194-2207. doi: 10.1038/s41418-019-0283-z. Epub 2019 Feb 13. Cell Death Differ. 2019. PMID: 30760872 Free PMC article.
-
SUMO and Chromatin Remodeling.Adv Exp Med Biol. 2017;963:35-50. doi: 10.1007/978-3-319-50044-7_3. Adv Exp Med Biol. 2017. PMID: 28197905 Review.
Cited by
-
Mammalian PRC1 Complexes: Compositional Complexity and Diverse Molecular Mechanisms.Int J Mol Sci. 2020 Nov 14;21(22):8594. doi: 10.3390/ijms21228594. Int J Mol Sci. 2020. PMID: 33202645 Free PMC article. Review.
-
MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6.PLoS Genet. 2018 Jan 30;14(1):e1007193. doi: 10.1371/journal.pgen.1007193. eCollection 2018 Jan. PLoS Genet. 2018. PMID: 29381691 Free PMC article.
-
ADP-ribosyltransferases Parp1 and Parp7 safeguard pluripotency of ES cells.Nucleic Acids Res. 2014 Aug;42(14):8914-27. doi: 10.1093/nar/gku591. Epub 2014 Jul 17. Nucleic Acids Res. 2014. PMID: 25034692 Free PMC article.
-
SUMOylation regulates cytochrome P450 2E1 expression and activity in alcoholic liver disease.FASEB J. 2018 Jun;32(6):3278-3288. doi: 10.1096/fj.201701124R. Epub 2018 Jan 18. FASEB J. 2018. PMID: 29401608 Free PMC article.
-
SUMOylated non-canonical polycomb PRC1.6 complex as a prerequisite for recruitment of transcription factor RBPJ.Epigenetics Chromatin. 2021 Jul 31;14(1):38. doi: 10.1186/s13072-021-00412-9. Epigenetics Chromatin. 2021. PMID: 34332624 Free PMC article.
References
-
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 2003;28:69–74. - PubMed
-
- Santiveri CM, Lechtenberg BC, Allen MD, Sathyamurthy A, Jaulent AM, Freund SM, Bycroft M. The malignant brain tumor repeats of human SCML2 bind to peptides containing monomethylated lysine. J. Mol. Biol. 2008;382:1107–1112. - PubMed
-
- Min J, Allali-Hassani A, Nady N, Qi C, Ouyang H, Liu Y, MacKenzie F, Vedadi M, Arrowsmith CH. L3MBTL1 recognition of mono- and dimethylated histones. Nature Struct. Mol. Biol. 2007;14:1229–1230. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

