HIV-1-associated PKA acts as a cofactor for genome reverse transcription
- PMID: 24344931
- PMCID: PMC3880072
- DOI: 10.1186/1742-4690-10-157
HIV-1-associated PKA acts as a cofactor for genome reverse transcription
Abstract
Background: Host cell proteins, including cellular kinases, are embarked into intact HIV-1 particles. We have previously shown that the Cα catalytic subunit of cAMP-dependent protein kinase is packaged within HIV-1 virions as an enzymatically active form able to phosphorylate a synthetic substrate in vitro (Cartier et al. J. Biol. Chem. 278:35211 (2003)). The present study was conceived to investigate the contribution of HIV-1-associated PKA to the retroviral life cycle.
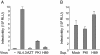
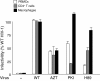
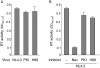
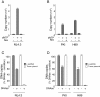
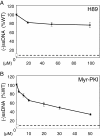
Results: NL4.3 viruses were produced from cells cultured in the presence of PKA inhibitors H89 (H89-NL4.3) or Myr-PKI (PKI-NL4.3) and analyzed for viral replication. Despite being mature and normally assembled, and containing expected levels of genomic RNA and RT enzymatic activity, such viruses showed poor infectivity. Indeed, infection generated reduced amounts of strong-strop minus strand DNA, while incoming RNA levels in target cells were unaffected. Decreased cDNA synthesis was also evidenced in intact H89-NL4.3 and PKI-NL4.3 cell free particles using endogenous reverse transcription (ERT) experiments. Moreover, similar defects were reproduced when wild type NL4.3 particles preincubated with PKA inhibitors were subjected to ERT reactions.
Conclusions: Altogether, our results indicate that HIV-1-associated PKA is required for early reverse transcription of the retroviral genome both in cell free intact viruses and in target cells. Accordingly, virus-associated PKA behaves as a cofactor of an intraviral process required for optimal reverse transcription and for early post-entry events.
Figures

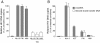
Similar articles
-
Active cAMP-dependent protein kinase incorporated within highly purified HIV-1 particles is required for viral infectivity and interacts with viral capsid protein.J Biol Chem. 2003 Sep 12;278(37):35211-9. doi: 10.1074/jbc.M301257200. Epub 2003 Jul 3. J Biol Chem. 2003. PMID: 12842892
-
INI1/hSNF5-interaction defective HIV-1 IN mutants exhibit impaired particle morphology, reverse transcription and integration in vivo.Retrovirology. 2013 Jun 24;10:66. doi: 10.1186/1742-4690-10-66. Retrovirology. 2013. PMID: 23799881 Free PMC article.
-
Host SAMHD1 protein restricts endogenous reverse transcription of HIV-1 in nondividing macrophages.Retrovirology. 2018 Oct 13;15(1):69. doi: 10.1186/s12977-018-0452-z. Retrovirology. 2018. PMID: 30316304 Free PMC article.
-
HIV-1 uncoating: connection to nuclear entry and regulation by host proteins.Virology. 2014 Apr;454-455:371-9. doi: 10.1016/j.virol.2014.02.004. Epub 2014 Feb 20. Virology. 2014. PMID: 24559861 Free PMC article. Review.
-
Strand transfer events during HIV-1 reverse transcription.Virus Res. 2008 Jun;134(1-2):19-38. doi: 10.1016/j.virusres.2007.12.017. Epub 2008 Feb 14. Virus Res. 2008. PMID: 18279992 Review.
Cited by
-
Cellular minichromosome maintenance complex component 5 (MCM5) is incorporated into HIV-1 virions and modulates viral replication in the newly infected cells.Virology. 2016 Oct;497:11-22. doi: 10.1016/j.virol.2016.06.023. Epub 2016 Jul 12. Virology. 2016. PMID: 27414250 Free PMC article.
-
Increased cAMP-PKA signaling pathway activation is involved in up-regulation of CTLA-4 expression in CD4+ T cells in acute SIVmac239-infected Chinese rhesus macaques.Virus Res. 2024 Mar;341:199313. doi: 10.1016/j.virusres.2024.199313. Epub 2024 Jan 22. Virus Res. 2024. PMID: 38244614 Free PMC article.
-
Phosphorylation of the HIV-1 capsid by MELK triggers uncoating to promote viral cDNA synthesis.PLoS Pathog. 2017 Jul 6;13(7):e1006441. doi: 10.1371/journal.ppat.1006441. eCollection 2017 Jul. PLoS Pathog. 2017. PMID: 28683086 Free PMC article.
-
The PKA-CREB1 axis regulates coronavirus proliferation by viral helicase nsp13 association.J Virol. 2024 Apr 16;98(4):e0156523. doi: 10.1128/jvi.01565-23. Epub 2024 Mar 6. J Virol. 2024. PMID: 38445884 Free PMC article.
-
Does BCA3 Play a Role in the HIV-1 Replication Cycle?Viruses. 2018 Apr 20;10(4):212. doi: 10.3390/v10040212. Viruses. 2018. PMID: 29677171 Free PMC article.
References
-
- Abbink TE, Berkhout B. HIV-1 reverse transcription initiation: a potential target for novel antivirals? Virus Res. 2008;10:4–18. - PubMed
-
- Darlix JL, Godet J, Ivanyi-Nagy R, Fosse P, Mauffret O, Mely Y. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J Mol Biol. 2011;10:565–581. - PubMed
-
- Gleenberg IO, Herschhorn A, Hizi A. Inhibition of the activities of reverse transcriptase and integrase of human immunodeficiency virus type-1 by peptides derived from the homologous viral protein R (Vpr) J Mol Biol. 2007;10:1230–1243. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

