Development of a novel DNA-launched dengue virus type 2 infectious clone assembled in a bacterial artificial chromosome
- PMID: 24342140
- PMCID: PMC7114509
- DOI: 10.1016/j.virusres.2013.12.001
Development of a novel DNA-launched dengue virus type 2 infectious clone assembled in a bacterial artificial chromosome
Abstract
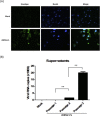
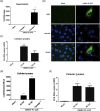
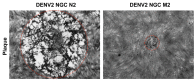
Major progress in Dengue virus (DENV) biology has resulted from the use of infectious clones obtained through reverse genetics. The construction of these clones is commonly based on high- or low-copy number plasmids, yeast artificial chromosomes, yeast-Escherichia coli shuttle vectors, and bacterial artificial chromosomes (BACs). Prokaryotic promoters have consistently been used for the transcription of these clones. The goal of this study was to develop a novel DENV infectious clone in a BAC under the control of the cytomegalovirus immediate-early promoter and to generate a virus with the fusion envelope-green fluorescent protein in an attempt to track virus infection. The transfection of Vero cells with a plasmid encoding the DENV infectious clone facilitated the recovery of infectious particles that increased in titer after serial passages in C6/36 cells. The plaque size and syncytia phenotypes of the recombinant virus were similar to those of the parental virus. Despite the observation of autonomous replication and the detection of low levels of viral genome after two passages, the insertion of green fluorescent protein and Renilla luciferase reporter genes negatively impacted virus rescue. To the best of our knowledge, this is the first study using a DENV infectious clone under the control of the cytomegalovirus promoter to facilitate the recovery of recombinant viruses without the need for in vitro transcription. This novel molecular clone will be useful for establishing the molecular basis of replication, assembly, and pathogenesis, evaluating potential antiviral drugs, and the development of vaccine candidates for attenuated recombinant viruses.
Keywords: Bacterial artificial chromosome; Dengue virus; Eukaryotic promoter; Flavivirus; Infectious cDNA; Reverse genetics.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures




Similar articles
-
Identification of an Arylnaphthalene Lignan Derivative as an Inhibitor against Dengue Virus Serotypes 1 to 4 (DENV-1 to -4) Using a Newly Developed DENV-3 Infectious Clone and Replicon.Microbiol Spectr. 2023 Aug 17;11(4):e0042323. doi: 10.1128/spectrum.00423-23. Epub 2023 Jun 28. Microbiol Spectr. 2023. PMID: 37378517 Free PMC article.
-
Successful propagation of flavivirus infectious cDNAs by a novel method to reduce the cryptic bacterial promoter activity of virus genomes.J Virol. 2011 Mar;85(6):2927-41. doi: 10.1128/JVI.01986-10. Epub 2011 Jan 12. J Virol. 2011. PMID: 21228244 Free PMC article.
-
Construction and characterisation of a complete reverse genetics system of dengue virus type 3.Mem Inst Oswaldo Cruz. 2013 Dec;108(8):983-91. doi: 10.1590/0074-0276130298. Mem Inst Oswaldo Cruz. 2013. PMID: 24402142 Free PMC article.
-
Establishment and Application of Flavivirus Replicons.Adv Exp Med Biol. 2018;1062:165-173. doi: 10.1007/978-981-10-8727-1_12. Adv Exp Med Biol. 2018. PMID: 29845532 Review.
-
Dengue Virus Reporter Replicon is a Valuable Tool for Antiviral Drug Discovery and Analysis of Virus Replication Mechanisms.Viruses. 2016 May 5;8(5):122. doi: 10.3390/v8050122. Viruses. 2016. PMID: 27164125 Free PMC article. Review.
Cited by
-
Full-length infectious clone of a low passage dengue virus serotype 2 from Brazil.Mem Inst Oswaldo Cruz. 2015 Aug;110(5):677-83. doi: 10.1590/0074-02760150053. Epub 2015 Jul 17. Mem Inst Oswaldo Cruz. 2015. PMID: 26200712 Free PMC article.
-
Enterovirus A71 DNA-Launched Infectious Clone as a Robust Reverse Genetic Tool.PLoS One. 2016 Sep 12;11(9):e0162771. doi: 10.1371/journal.pone.0162771. eCollection 2016. PLoS One. 2016. PMID: 27617744 Free PMC article.
-
Bacterial Artificial Chromosomes: A Functional Genomics Tool for the Study of Positive-strand RNA Viruses.J Vis Exp. 2015 Dec 29;(106):e53164. doi: 10.3791/53164. J Vis Exp. 2015. PMID: 26780115 Free PMC article.
-
Epitope Addition and Ablation via Manipulation of a Dengue Virus Serotype 1 Infectious Clone.mSphere. 2017 Feb 22;2(1):e00380-16. doi: 10.1128/mSphere.00380-16. eCollection 2017 Jan-Feb. mSphere. 2017. PMID: 28251184 Free PMC article.
-
Construction and characterization of an improved DNA-launched infectious clone of duck hepatitis a virus type 1.Virol J. 2017 Nov 3;14(1):212. doi: 10.1186/s12985-017-0883-5. Virol J. 2017. PMID: 29100535 Free PMC article.
References
-
- Bartenschlager R., Miller S. Molecular aspects of dengue virus replication. Future Microbiology. 2008;3(2):155–165. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

