High-resolution mapping of transcription factor binding sites on native chromatin
- PMID: 24336359
- PMCID: PMC3929178
- DOI: 10.1038/nmeth.2766
High-resolution mapping of transcription factor binding sites on native chromatin
Abstract
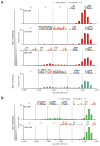
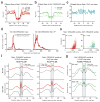
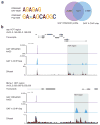
Sequence-specific DNA-binding proteins including transcription factors (TFs) are key determinants of gene regulation and chromatin architecture. TF profiling is commonly carried out by formaldehyde cross-linking and sonication followed by chromatin immunoprecipitation (X-ChIP). We describe a method to profile TF binding at high resolution without cross-linking. We begin with micrococcal nuclease-digested non-cross-linked chromatin and then perform affinity purification of TFs and paired-end sequencing. The resulting occupied regions of genomes from affinity-purified naturally isolated chromatin (ORGANIC) profiles of Saccharomyces cerevisiae Abf1 and Reb1 provide high-resolution maps that are accurate, as defined by the presence of known TF consensus motifs in identified binding sites, that are not biased toward accessible chromatin and that do not require input normalization. We profiled Drosophila melanogaster GAGA factor and Pipsqueak to test ORGANIC performance on larger genomes. Our results suggest that ORGANIC profiling is a widely applicable high-resolution method for sensitive and specific profiling of direct protein-DNA interactions.
Conflict of interest statement
None.
Figures



Similar articles
-
Mapping regulatory factors by immunoprecipitation from native chromatin.Curr Protoc Mol Biol. 2015 Apr 1;110:21.31.1-21.31.25. doi: 10.1002/0471142727.mb2131s110. Curr Protoc Mol Biol. 2015. PMID: 25827087 Free PMC article.
-
Different chromatin interfaces of the Drosophila dosage compensation complex revealed by high-shear ChIP-seq.Genome Res. 2013 Mar;23(3):473-85. doi: 10.1101/gr.146407.112. Epub 2012 Dec 11. Genome Res. 2013. PMID: 23233545 Free PMC article.
-
Genome-wide off-rates reveal how DNA binding dynamics shape transcription factor function.Mol Syst Biol. 2020 Oct;16(10):e9885. doi: 10.15252/msb.20209885. Mol Syst Biol. 2020. PMID: 33280256 Free PMC article.
-
Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo.Curr Protoc Cell Biol. 2004 Sep;Chapter 17:Unit 17.7. doi: 10.1002/0471143030.cb1707s23. Curr Protoc Cell Biol. 2004. PMID: 18228445 Review.
-
A conserved role for transcription factor sumoylation in binding-site selection.Curr Genet. 2019 Dec;65(6):1307-1312. doi: 10.1007/s00294-019-00992-w. Epub 2019 May 15. Curr Genet. 2019. PMID: 31093693 Review.
Cited by
-
Common genomic elements promote transcriptional and DNA replication roadblocks.Genome Res. 2016 Oct;26(10):1363-1375. doi: 10.1101/gr.204776.116. Epub 2016 Aug 18. Genome Res. 2016. PMID: 27540088 Free PMC article.
-
Active promoters give rise to false positive 'Phantom Peaks' in ChIP-seq experiments.Nucleic Acids Res. 2015 Aug 18;43(14):6959-68. doi: 10.1093/nar/gkv637. Epub 2015 Jun 27. Nucleic Acids Res. 2015. PMID: 26117547 Free PMC article.
-
Native internally calibrated chromatin immunoprecipitation for quantitative studies of histone post-translational modifications.Nat Protoc. 2019 Dec;14(12):3275-3302. doi: 10.1038/s41596-019-0218-7. Epub 2019 Nov 13. Nat Protoc. 2019. PMID: 31723301 Free PMC article.
-
RSC-Associated Subnucleosomes Define MNase-Sensitive Promoters in Yeast.Mol Cell. 2019 Jan 17;73(2):238-249.e3. doi: 10.1016/j.molcel.2018.10.046. Epub 2018 Dec 13. Mol Cell. 2019. PMID: 30554944 Free PMC article.
-
INO80 Chromatin Remodeling Coordinates Metabolic Homeostasis with Cell Division.Cell Rep. 2018 Jan 16;22(3):611-623. doi: 10.1016/j.celrep.2017.12.079. Cell Rep. 2018. PMID: 29346761 Free PMC article.
References
-
- Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. - PubMed
-
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. - PubMed
-
- O’Neill LP, Turner BM. Immunoprecipitation of native chromatin: NChIP. Methods. 2003;31:76–82. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

