Primary cilia are specialized calcium signalling organelles
- PMID: 24336288
- PMCID: PMC4112737
- DOI: 10.1038/nature12833
Primary cilia are specialized calcium signalling organelles
Abstract
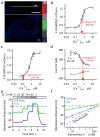
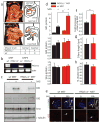
Primary cilia are solitary, non-motile extensions of the centriole found on nearly all nucleated eukaryotic cells between cell divisions. Only ∼200-300 nm in diameter and a few micrometres long, they are separated from the cytoplasm by the ciliary neck and basal body. Often called sensory cilia, they are thought to receive chemical and mechanical stimuli and initiate specific cellular signal transduction pathways. When activated by a ligand, hedgehog pathway proteins, such as GLI2 and smoothened (SMO), translocate from the cell into the cilium. Mutations in primary ciliary proteins are associated with severe developmental defects. The ionic conditions, permeability of the primary cilia membrane, and effectiveness of the diffusion barriers between the cilia and cell body are unknown. Here we show that cilia are a unique calcium compartment regulated by a heteromeric TRP channel, PKD1L1-PKD2L1, in mice and humans. In contrast to the hypothesis that polycystin (PKD) channels initiate changes in ciliary calcium that are conducted into the cytoplasm, we show that changes in ciliary calcium concentration occur without substantially altering global cytoplasmic calcium. PKD1L1-PKD2L1 acts as a ciliary calcium channel controlling ciliary calcium concentration and thereby modifying SMO-activated GLI2 translocation and GLI1 expression.
Figures
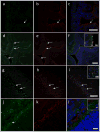
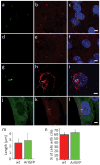
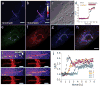
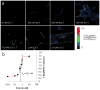
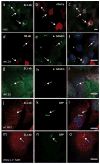
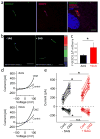
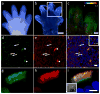
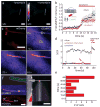
Comment in
-
Calcium: an ion channel for cilia.Nat Rev Mol Cell Biol. 2014 Feb;15(2):78. doi: 10.1038/nrm3736. Epub 2013 Dec 27. Nat Rev Mol Cell Biol. 2014. PMID: 24370826 No abstract available.
Similar articles
-
Direct recording and molecular identification of the calcium channel of primary cilia.Nature. 2013 Dec 12;504(7479):315-8. doi: 10.1038/nature12832. Nature. 2013. PMID: 24336289 Free PMC article.
-
Cell context-specific expression of primary cilia in the human testis and ciliary coordination of Hedgehog signalling in mouse Leydig cells.Sci Rep. 2015 May 20;5:10364. doi: 10.1038/srep10364. Sci Rep. 2015. PMID: 25992706 Free PMC article.
-
Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus.Proc Natl Acad Sci U S A. 2009 Dec 22;106(51):21666-71. doi: 10.1073/pnas.0912180106. Epub 2009 Dec 8. Proc Natl Acad Sci U S A. 2009. PMID: 19996169 Free PMC article.
-
Signaling in the primary cilium through the lens of the Hedgehog pathway.Wiley Interdiscip Rev Dev Biol. 2020 Nov;9(6):e377. doi: 10.1002/wdev.377. Epub 2020 Feb 21. Wiley Interdiscip Rev Dev Biol. 2020. PMID: 32084300 Free PMC article. Review.
-
The primary cilia, a 'Rab-id' transit system for hedgehog signaling.Curr Opin Cell Biol. 2007 Dec;19(6):691-6. doi: 10.1016/j.ceb.2007.10.008. Epub 2007 Dec 3. Curr Opin Cell Biol. 2007. PMID: 18061425 Free PMC article. Review.
Cited by
-
Fluorescence imaging of beta cell primary cilia.Front Endocrinol (Lausanne). 2022 Sep 23;13:1004136. doi: 10.3389/fendo.2022.1004136. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36213262 Free PMC article.
-
Systematic Discovery of Short Linear Motifs Decodes Calcineurin Phosphatase Signaling.Mol Cell. 2020 Jul 16;79(2):342-358.e12. doi: 10.1016/j.molcel.2020.06.029. Epub 2020 Jul 8. Mol Cell. 2020. PMID: 32645368 Free PMC article.
-
Nanobody-directed targeting of optogenetic tools to study signaling in the primary cilium.Elife. 2020 Jun 24;9:e57907. doi: 10.7554/eLife.57907. Elife. 2020. PMID: 32579112 Free PMC article.
-
Mutations in human C2CD3 cause skeletal dysplasia and provide new insights into phenotypic and cellular consequences of altered C2CD3 function.Sci Rep. 2016 Apr 20;6:24083. doi: 10.1038/srep24083. Sci Rep. 2016. PMID: 27094867 Free PMC article.
-
Role of Ca2+ transients at the node of the mouse embryo in breaking of left-right symmetry.Sci Adv. 2020 Jul 22;6(30):eaba1195. doi: 10.1126/sciadv.aba1195. eCollection 2020 Jul. Sci Adv. 2020. PMID: 32743070 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

