Assessment of viral community functional potential from viral metagenomes may be hampered by contamination with cellular sequences
- PMID: 24335607
- PMCID: PMC3877843
- DOI: 10.1098/rsob.130160
Assessment of viral community functional potential from viral metagenomes may be hampered by contamination with cellular sequences
Abstract
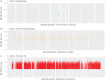
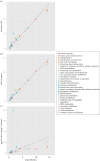
Although the importance of viruses in natural ecosystems is widely acknowledged, the functional potential of viral communities is yet to be determined. Viral genomes are traditionally believed to carry only those genes that are directly pertinent to the viral life cycle, though this view was challenged by the discovery of metabolism genes in several phage genomes. Metagenomic approaches extended these analyses to a community scale, and several studies concluded that microbial and viral communities encompass similar functional potentials. However, these conclusions could originate from the presence of cellular DNA within viral metagenomes. We developed a computational method to estimate the proportion and origin of cellular sequences in a set of 67 published viromes. A quarter of the datasets were found to contain a substantial amount of sequences originating from cellular genomes. When considering only viromes with no cellular DNA detected, the functional potential of viral and microbial communities was found to be fundamentally different-a conclusion more consistent with the actual picture drawn from known viruses. Yet a significant number of cellular metabolism genes was still retrieved in these viromes, suggesting that the presence of auxiliary genes involved in various metabolic pathways within viral genomes is a general trend in the virosphere.
Keywords: functional potential; metagenomics; phages; viruses.
Figures


Similar articles
-
Viromes vs. mixed community metagenomes: choice of method dictates interpretation of viral community ecology.Microbiome. 2024 Oct 7;12(1):195. doi: 10.1186/s40168-024-01905-x. Microbiome. 2024. PMID: 39375774 Free PMC article.
-
Antibiotics in feed induce prophages in swine fecal microbiomes.mBio. 2011 Nov 29;2(6):e00260-11. doi: 10.1128/mBio.00260-11. Print 2011. mBio. 2011. PMID: 22128350 Free PMC article.
-
Phage hunters: Computational strategies for finding phages in large-scale 'omics datasets.Virus Res. 2018 Jan 15;244:110-115. doi: 10.1016/j.virusres.2017.10.019. Epub 2017 Nov 1. Virus Res. 2018. PMID: 29100906 Review.
-
Microbial Diversity and Phage-Host Interactions in the Georgian Coastal Area of the Black Sea Revealed by Whole Genome Metagenomic Sequencing.Mar Drugs. 2020 Nov 14;18(11):558. doi: 10.3390/md18110558. Mar Drugs. 2020. PMID: 33202695 Free PMC article.
-
Computational prospecting the great viral unknown.FEMS Microbiol Lett. 2016 May;363(10):fnw077. doi: 10.1093/femsle/fnw077. Epub 2016 Mar 29. FEMS Microbiol Lett. 2016. PMID: 27030726 Review.
Cited by
-
Insights into the global freshwater virome.Front Microbiol. 2022 Sep 28;13:953500. doi: 10.3389/fmicb.2022.953500. eCollection 2022. Front Microbiol. 2022. PMID: 36246212 Free PMC article.
-
RNA Viruses in Aquatic Ecosystems through the Lens of Ecological Genomics and Transcriptomics.Viruses. 2022 Mar 28;14(4):702. doi: 10.3390/v14040702. Viruses. 2022. PMID: 35458432 Free PMC article. Review.
-
Murine colitis reveals a disease-associated bacteriophage community.Nat Microbiol. 2018 Sep;3(9):1023-1031. doi: 10.1038/s41564-018-0210-y. Epub 2018 Jul 23. Nat Microbiol. 2018. PMID: 30038310 Free PMC article.
-
Dietary energy drives the dynamic response of bovine rumen viral communities.Microbiome. 2017 Nov 28;5(1):155. doi: 10.1186/s40168-017-0374-3. Microbiome. 2017. PMID: 29179741 Free PMC article.
-
Phages of life - the path to pharma.Br J Pharmacol. 2018 Feb;175(3):412-418. doi: 10.1111/bph.14106. Epub 2018 Jan 3. Br J Pharmacol. 2018. PMID: 29266197 Free PMC article. Review.
References
-
- Bergh O, Børsheim KY, Bratbak G, Heldal M. 1989. High abundance of viruses found in aquatic environments. Nature 340, 467–468 (doi:10.1038/340467a0) - DOI - PubMed
-
- Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (doi:10.1038/nrmicro1750) - DOI - PubMed
-
- Wommack KE, Colwell RR. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64, 69–114 (doi:10.1128/MMBR.64.1.69-114.2000) - DOI - PMC - PubMed
-
- Rodriguez-Valera F, Martin-Cuadrado A-B, Rodriguez-Brito B, Pasić L, Thingstad TF, Rohwer F, Mira A. 2009. Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7, 828–836 (doi:10.1038/nrmicro2235) - DOI - PubMed
-
- Forterre P. 2006. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 117, 5–16 (doi:10.1016/j.virusres.2006.01.010) - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

