The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns
- PMID: 24316672
- PMCID: PMC4019675
- DOI: 10.1038/ncb2885
The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns
Abstract
Bacterial cytokinesis is commonly initiated by the Z-ring, a cytoskeletal structure that assembles at the site of division. Its primary component is FtsZ, a tubulin superfamily GTPase, which is recruited to the membrane by the actin-related protein FtsA. Both proteins are required for the formation of the Z-ring, but if and how they influence each other's assembly dynamics is not known. Here, we reconstituted FtsA-dependent recruitment of FtsZ polymers to supported membranes, where both proteins self-organize into complex patterns, such as fast-moving filament bundles and chirally rotating rings. Using fluorescence microscopy and biochemical perturbations, we found that these large-scale rearrangements of FtsZ emerge from its polymerization dynamics and a dual, antagonistic role of FtsA: recruitment of FtsZ filaments to the membrane and negative regulation of FtsZ organization. Our findings provide a model for the initial steps of bacterial cell division and illustrate how dynamic polymers can self-organize into large-scale structures.
Figures


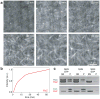
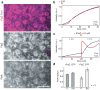
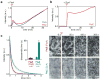

Comment in
-
Bacterial physiology: remodelling the FtsZ network.Nat Rev Microbiol. 2014 Feb;12(2):77. doi: 10.1038/nrmicro3196. Epub 2013 Dec 16. Nat Rev Microbiol. 2014. PMID: 24336149 No abstract available.
-
Cytoskeleton: Remodelling the FtsZ network.Nat Rev Mol Cell Biol. 2014 Jan;15(1):3. doi: 10.1038/nrm3724. Nat Rev Mol Cell Biol. 2014. PMID: 24355980 No abstract available.
-
Bacterial cell division: a swirling ring to rule them all?Curr Biol. 2014 Feb 17;24(4):R157-9. doi: 10.1016/j.cub.2014.01.025. Curr Biol. 2014. PMID: 24556438
Similar articles
-
Bacterial cell division: a swirling ring to rule them all?Curr Biol. 2014 Feb 17;24(4):R157-9. doi: 10.1016/j.cub.2014.01.025. Curr Biol. 2014. PMID: 24556438
-
Assembly and architecture of Escherichia coli divisome proteins FtsA and FtsZ.J Biol Chem. 2022 Mar;298(3):101663. doi: 10.1016/j.jbc.2022.101663. Epub 2022 Jan 29. J Biol Chem. 2022. PMID: 35104502 Free PMC article.
-
Escherichia coli ZipA Organizes FtsZ Polymers into Dynamic Ring-Like Protofilament Structures.mBio. 2018 Jun 19;9(3):e01008-18. doi: 10.1128/mBio.01008-18. mBio. 2018. PMID: 29921670 Free PMC article.
-
The keepers of the ring: regulators of FtsZ assembly.FEMS Microbiol Rev. 2016 Jan;40(1):57-67. doi: 10.1093/femsre/fuv040. Epub 2015 Sep 15. FEMS Microbiol Rev. 2016. PMID: 26377318 Review.
-
FtsZ dynamics in bacterial division: What, how, and why?Curr Opin Cell Biol. 2021 Feb;68:163-172. doi: 10.1016/j.ceb.2020.10.013. Epub 2020 Nov 18. Curr Opin Cell Biol. 2021. PMID: 33220539 Free PMC article. Review.
Cited by
-
Chiral and nematic phases of flexible active filaments.Nat Phys. 2023;19(12):1916-1926. doi: 10.1038/s41567-023-02218-w. Epub 2023 Oct 9. Nat Phys. 2023. PMID: 38075437 Free PMC article.
-
Enhanced production of polyhydroxybutyrate by multiple dividing E. coli.Microb Cell Fact. 2016 Jul 27;15(1):128. doi: 10.1186/s12934-016-0531-6. Microb Cell Fact. 2016. PMID: 27465264 Free PMC article.
-
Complex state transitions of the bacterial cell division protein FtsZ.Mol Biol Cell. 2024 Oct 1;35(10):ar130. doi: 10.1091/mbc.E23-11-0446. Epub 2024 Jul 31. Mol Biol Cell. 2024. PMID: 39083352 Free PMC article.
-
Phosphorylation of FtsZ and FtsA by a DNA Damage-Responsive Ser/Thr Protein Kinase Affects Their Functional Interactions in Deinococcus radiodurans.mSphere. 2018 Jul 18;3(4):e00325-18. doi: 10.1128/mSphere.00325-18. mSphere. 2018. PMID: 30021877 Free PMC article.
-
Cell cycle-dependent regulation of FtsZ in Escherichia coli in slow growth conditions.Mol Microbiol. 2018 Dec;110(6):1030-1044. doi: 10.1111/mmi.14135. Epub 2018 Oct 29. Mol Microbiol. 2018. PMID: 30230648 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

