Protein arginine methylation of non-histone proteins and its role in diseases
- PMID: 24296620
- PMCID: PMC3925732
- DOI: 10.4161/cc.27353
Protein arginine methylation of non-histone proteins and its role in diseases
Abstract
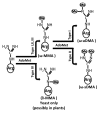
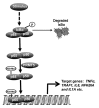
Protein arginine methyltransferases (PRMTs) are a family of enzymes that can methylate arginine residues on histones and other proteins. PRMTs play a crucial role in influencing various cellular functions, including cellular development and tumorigenesis. Arginine methylation by PRMTs is found on both nuclear and cytoplasmic proteins. Recently, there is increasing evidence regarding post-translational modifications of non-histone proteins by PRMTs, illustrating the previously unknown importance of PRMTs in the regulation of various cellular functions by post-translational modifications. In this review, we present the recent developments in the regulation of non-histone proteins by PRMTs.
Keywords: arginine; post-translational modification; protein arginine methyltransferases.
Figures

Similar articles
-
Protein arginine methyltransferases (PRMTs): role in chromatin organization.Adv Biol Regul. 2015 Jan;57:173-84. doi: 10.1016/j.jbior.2014.09.003. Epub 2014 Sep 17. Adv Biol Regul. 2015. PMID: 25263650 Review.
-
Research progress on PRMTs involved in epigenetic modification and tumour signalling pathway regulation (Review).Int J Oncol. 2023 May;62(5):62. doi: 10.3892/ijo.2023.5510. Epub 2023 Apr 7. Int J Oncol. 2023. PMID: 37026519 Free PMC article. Review.
-
Non-Histone Arginine Methylation by Protein Arginine Methyltransferases.Curr Protein Pept Sci. 2020;21(7):699-712. doi: 10.2174/1389203721666200507091952. Curr Protein Pept Sci. 2020. PMID: 32379587 Free PMC article. Review.
-
Assaying epigenome functions of PRMTs and their substrates.Methods. 2020 Mar 15;175:53-65. doi: 10.1016/j.ymeth.2019.09.014. Epub 2019 Sep 19. Methods. 2020. PMID: 31542509 Review.
-
The regulation, functions and clinical relevance of arginine methylation.Nat Rev Mol Cell Biol. 2019 Oct;20(10):642-657. doi: 10.1038/s41580-019-0155-x. Epub 2019 Jul 26. Nat Rev Mol Cell Biol. 2019. PMID: 31350521 Review.
Cited by
-
Dysregulation of histone methyltransferases in breast cancer - Opportunities for new targeted therapies?Mol Oncol. 2016 Dec;10(10):1497-1515. doi: 10.1016/j.molonc.2016.09.003. Epub 2016 Sep 23. Mol Oncol. 2016. PMID: 27717710 Free PMC article. Review.
-
Arginine Methyltransferase PRMT8 Provides Cellular Stress Tolerance in Aging Motoneurons.J Neurosci. 2018 Aug 29;38(35):7683-7700. doi: 10.1523/JNEUROSCI.3389-17.2018. Epub 2018 Jul 27. J Neurosci. 2018. PMID: 30054395 Free PMC article.
-
Islet-specific Prmt5 excision leads to reduced insulin expression and glucose intolerance in mice.J Endocrinol. 2020 Jan 1;244(1):41-52. doi: 10.1530/JOE-19-0268. J Endocrinol. 2020. PMID: 31539871 Free PMC article.
-
NF-κB: Regulation by Methylation.Cancer Res. 2015 Sep 15;75(18):3692-5. doi: 10.1158/0008-5472.CAN-15-1022. Epub 2015 Sep 3. Cancer Res. 2015. PMID: 26337909 Free PMC article. Review.
-
Identification of hnRNP-A1 as a pharmacodynamic biomarker of type I PRMT inhibition in blood and tumor tissues.Sci Rep. 2020 Dec 17;10(1):22155. doi: 10.1038/s41598-020-78800-6. Sci Rep. 2020. PMID: 33335114 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
