Mechanism of human PTEN localization revealed by heterologous expression in Dictyostelium
- PMID: 24292679
- PMCID: PMC4041858
- DOI: 10.1038/onc.2013.507
Mechanism of human PTEN localization revealed by heterologous expression in Dictyostelium
Abstract
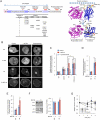
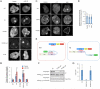
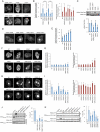
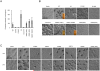
Phosphatase and tensin homolog (PTEN) is one of the most frequently mutated tumor suppressor genes in cancers. PTEN has a central role in phosphatidylinositol (3,4,5)-trisphosphate (PIP3) signaling and converts PIP3 to phosphatidylinositol (4,5)-bisphosphate at the plasma membrane. Despite its importance, the mechanism that mediates membrane localization of PTEN is poorly understood. Here, we generated a library that contains green fluorescent protein fused to randomly mutated human PTEN and expressed the library in Dictyostelium cells. Using live cell imaging, we identified mutations that enhance the association of PTEN with the plasma membrane. These mutations were located in four separate regions, including the phosphatase catalytic site, the calcium-binding region 3 (CBR3) loop, the Cα2 loop and the C-terminal tail phosphorylation site. The phosphatase catalytic site, the CBR3 loop and the Cα2 loop formed the membrane-binding regulatory interface and interacted with the inhibitory phosphorylated C-terminal tail. Furthermore, we showed that membrane recruitment of PTEN is required for PTEN function in cells. Thus, heterologous expression system in Dictyostelium cells provides mechanistic and functional insight into membrane localization of PTEN.
Figures

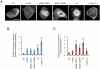
Similar articles
-
Engineering PTEN function: membrane association and activity.Methods. 2015 May;77-78:119-24. doi: 10.1016/j.ymeth.2014.10.018. Epub 2014 Oct 22. Methods. 2015. PMID: 25448479 Free PMC article.
-
Engineering ePTEN, an enhanced PTEN with increased tumor suppressor activities.Proc Natl Acad Sci U S A. 2014 Jul 1;111(26):E2684-93. doi: 10.1073/pnas.1409433111. Epub 2014 Jun 16. Proc Natl Acad Sci U S A. 2014. PMID: 24979808 Free PMC article.
-
A new class of cancer-associated PTEN mutations defined by membrane translocation defects.Oncogene. 2015 Jul;34(28):3737-43. doi: 10.1038/onc.2014.293. Epub 2014 Sep 29. Oncogene. 2015. PMID: 25263454 Free PMC article.
-
Structural Mechanisms of PTEN Regulation.Cold Spring Harb Perspect Med. 2020 Mar 2;10(3):a036152. doi: 10.1101/cshperspect.a036152. Cold Spring Harb Perspect Med. 2020. PMID: 31636093 Free PMC article. Review.
-
Regulation of PTEN function as a PIP3 gatekeeper through membrane interaction.Cell Cycle. 2006 Jul;5(14):1523-7. doi: 10.4161/cc.5.14.3005. Epub 2006 Jul 17. Cell Cycle. 2006. PMID: 16861931 Review.
Cited by
-
Fine-Tuning of Pten Localization and Phosphatase Activity Is Essential for Zebrafish Angiogenesis.PLoS One. 2016 May 3;11(5):e0154771. doi: 10.1371/journal.pone.0154771. eCollection 2016. PLoS One. 2016. PMID: 27138341 Free PMC article.
-
A minimal computational model for three-dimensional cell migration.J R Soc Interface. 2019 Dec;16(161):20190619. doi: 10.1098/rsif.2019.0619. Epub 2019 Dec 18. J R Soc Interface. 2019. PMID: 31847757 Free PMC article.
-
Structural and Dynamic Effects of PTEN C-Terminal Tail Phosphorylation.J Chem Inf Model. 2022 Sep 12;62(17):4175-4190. doi: 10.1021/acs.jcim.2c00441. Epub 2022 Aug 24. J Chem Inf Model. 2022. PMID: 36001481 Free PMC article.
-
Immunofluorescence Detection of Plasma Membranous PTEN in Cultured Cells.J Histochem Cytochem. 2022 Apr;70(4):289-297. doi: 10.1369/00221554221082539. Epub 2022 Feb 24. J Histochem Cytochem. 2022. PMID: 35199573 Free PMC article.
-
Multifaceted Regulation of PTEN Subcellular Distributions and Biological Functions.Cancers (Basel). 2019 Aug 26;11(9):1247. doi: 10.3390/cancers11091247. Cancers (Basel). 2019. PMID: 31454965 Free PMC article. Review.
References
-
- Leslie NR, Dixon MJ, Schenning M, Gray A, Batty IH. Distinct inactivation of PI3K signalling by PTEN and 5-phosphatases. Adv Biol Regul. 2012;52(1):205–13. Epub 2011/09/21. - PubMed
-
- Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol. 2013;10(3):143–53. Epub 2013/02/13. - PubMed
-
- Vanhaesebroeck B, Stephens L, Hawkins P. PI3K signalling: the path to discovery and understanding. Nature reviews Molecular cell biology. 2012;13(3):195–203. Epub 2012/02/24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

