Development and evaluation of a SYBR green-based real time RT-PCR assay for detection of the emerging avian influenza A (H7N9) virus
- PMID: 24278234
- PMCID: PMC3835827
- DOI: 10.1371/journal.pone.0080028
Development and evaluation of a SYBR green-based real time RT-PCR assay for detection of the emerging avian influenza A (H7N9) virus
Abstract
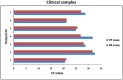
Most recently a novel avian-origin influenza A (H7N9) virus emerged in China and has been associated with lots of human infection and fatal cases. Genetic analysis of the viral genome revealed that this reassortant virus might be better adapted to humans than other avian influenza viruses. Molecular diagnostic methods are thus urgently needed in public health laboratories. In this study, a SYBR green-based one-step real time reverse transcription-PCR (RT-PCR) was developed to detect the novel H7N9 virus. The primer pairs on the basis of the hemagglutinin and neuraminidase gene sequences of H7N9 viruses amplified subtype-specific fragments with Tm values of 80.77±0.06°C for H7 and 81.20±0.17°C for N9 respectively. The standard curves showed a dynamic linear range across 6 log units of RNA copy number (10(6) to 10(1) copies/ µl) with a detection limit of 10 copies per reaction for both H7 and N9 assays by using serial ten-fold diluted in-vitro transcribed viral RNA. In addition, no cross-reactivity was observed with seasonal H1N1, H1N1 pdm09, H3N2, H5N1 and H9N2 viruses as well as other human respiratory viruses. When the assay was further evaluated in H7N9 virus infected clinical samples, positive amplification signals were obtained in all of the specimens with the accordance between H7 and N9 assays. Therefore, the established SYBR green-based real time RT-PCR assay could provide a rapid, sensitive, specific and reliable alternative approach with lower costs for high throughput screening of suspected samples from humans, animals and environments in first line public health laboratories.
Conflict of interest statement
Figures



Similar articles
-
Detection of avian influenza A/H7N9/2013 virus by real-time reverse transcription-polymerase chain reaction.J Virol Methods. 2014 Sep;206:140-3. doi: 10.1016/j.jviromet.2014.01.026. Epub 2014 May 29. J Virol Methods. 2014. PMID: 24880072
-
Development and evaluation of a real-time RT-PCR assay for detection of a novel avian influenza A (H5N6) virus.J Virol Methods. 2018 Jul;257:79-84. doi: 10.1016/j.jviromet.2018.05.001. Epub 2018 May 2. J Virol Methods. 2018. PMID: 29729298
-
Evaluation and application of a one-step duplex real-time reverse transcription polymerase chain reaction assay for the rapid detection of influenza A (H7N9) virus from poultry samples.Arch Virol. 2015 Oct;160(10):2471-7. doi: 10.1007/s00705-015-2511-2. Epub 2015 Jul 16. Arch Virol. 2015. PMID: 26179621
-
Rapid and sensitive detection of novel avian-origin influenza A (H7N9) virus by reverse transcription loop-mediated isothermal amplification combined with a lateral-flow device.PLoS One. 2013 Aug 1;8(8):e69941. doi: 10.1371/journal.pone.0069941. Print 2013. PLoS One. 2013. PMID: 23936359 Free PMC article.
-
Development of novel AllGlo-probe-based one-step multiplex qRT-PCR assay for rapid identification of avian influenza virus H7N9.Arch Virol. 2014 Jul;159(7):1707-13. doi: 10.1007/s00705-014-1979-5. Epub 2014 Jan 29. Arch Virol. 2014. PMID: 24473706
Cited by
-
Development and Characterization of SYBR Green I Based RT-PCR Assay for Detection of Omsk Hemorrhagic Fever Virus.Virol Sin. 2021 Dec;36(6):1644-1647. doi: 10.1007/s12250-021-00389-5. Epub 2021 Jun 2. Virol Sin. 2021. PMID: 34076867 Free PMC article. No abstract available.
-
Efficient Detection of Pathogenic Leptospires Using 16S Ribosomal RNA.PLoS One. 2015 Jun 19;10(6):e0128913. doi: 10.1371/journal.pone.0128913. eCollection 2015. PLoS One. 2015. PMID: 26091292 Free PMC article.
-
Novel, In-House, SYBR Green Based One-Step rRT-PCR: Rapid and Accurate Diagnosis of Crimean-Congo Hemorrhagic Fever Virus in Suspected Patients From Iran.Jundishapur J Microbiol. 2016 Jan 2;9(1):e29246. doi: 10.5812/jjm.29246. eCollection 2016 Jan. Jundishapur J Microbiol. 2016. PMID: 27099688 Free PMC article.
-
Multi-Target Strategy for Pan/Foot-and-Mouth Disease Virus (FMDV) Detection: A Combination of Sequences Analysis, in Silico Predictions and Laboratory Diagnostic Evaluation.Front Vet Sci. 2018 Jul 12;5:160. doi: 10.3389/fvets.2018.00160. eCollection 2018. Front Vet Sci. 2018. PMID: 30050913 Free PMC article.
-
Evaluation of Commercial Diagnostic Assays for the Specific Detection of Avian Influenza A (H7N9) Virus RNA Using a Quality-Control Panel and Clinical Specimens in China.PLoS One. 2015 Sep 11;10(9):e0137862. doi: 10.1371/journal.pone.0137862. eCollection 2015. PLoS One. 2015. PMID: 26361351 Free PMC article.
References
-
- Gao R, Cao B, Hu Y, Feng Z, Wang D, et al. (2013) Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368: 1888–1897. - PubMed
-
- World Health Organization Number of confirmed human cases of avian influenza A (H7N9) reported to WHO, as of 30 May 2013. Available: http://www.who.int/influenza/human_animal_interface/influenza_h7n9/08_Re....
-
- Li Q, Zhou L, Zhou M, Chen Z, Li F, et al... (2013) Preliminary Report: Epidemiology of the Avian Influenza A (H7N9) Outbreak in China. N Engl J Med.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

