Insights into mRNA export-linked molecular mechanisms of human disease through a Gle1 structure-function analysis
- PMID: 24275432
- PMCID: PMC3932673
- DOI: 10.1016/j.jbior.2013.10.002
Insights into mRNA export-linked molecular mechanisms of human disease through a Gle1 structure-function analysis
Abstract
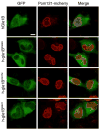
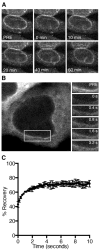
A critical step during gene expression is the directional export of nuclear messenger (m)RNA through nuclear pore complexes (NPCs) to the cytoplasm. During export, Gle1 in conjunction with inositol hexakisphosphate (IP6) spatially regulates the activity of the DEAD-box protein Dbp5 at the NPC cytoplasmic face. GLE1 mutations are causally linked to the human diseases lethal congenital contracture syndrome 1 (LCCS-1) and lethal arthrogryposis with anterior horn cell disease (LAAHD). Here, structure prediction and functional analysis provide strong evidence to suggest that the LCCS-1 and LAAHD disease mutations disrupt the function of Gle1 in mRNA export. Strikingly, direct fluorescence microscopy in living cells reveals a dramatic loss of steady-state NPC localization for GFP-gle1 proteins expressed from human gle1 genes harboring LAAHD and LCCS-1 mutations. The potential significance of these residues is further clarified by analyses of sequence and predicted structural conservation. This work offers insights into the perturbed mechanisms underlying human LCCS-1 and LAAHD disease states and emphasizes the potential impact of altered mRNA transport and gene expression in human disease.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
Survival beyond the perinatal period expands the phenotypes caused by mutations in GLE1.Am J Med Genet A. 2017 Nov;173(11):3098-3103. doi: 10.1002/ajmg.a.38406. Epub 2017 Sep 8. Am J Med Genet A. 2017. PMID: 28884921 Free PMC article.
-
Nup42 and IP6 coordinate Gle1 stimulation of Dbp5/DDX19B for mRNA export in yeast and human cells.Traffic. 2017 Dec;18(12):776-790. doi: 10.1111/tra.12526. Epub 2017 Oct 16. Traffic. 2017. PMID: 28869701 Free PMC article.
-
The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1.Genes Dev. 2011 May 15;25(10):1052-64. doi: 10.1101/gad.2041611. Genes Dev. 2011. PMID: 21576265 Free PMC article.
-
Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export.Nucleus. 2011 Nov-Dec;2(6):540-8. doi: 10.4161/nucl.2.6.17881. Epub 2011 Nov 1. Nucleus. 2011. PMID: 22064466 Free PMC article. Review.
-
Dbp5 - from nuclear export to translation.Biochim Biophys Acta. 2013 Aug;1829(8):791-8. doi: 10.1016/j.bbagrm.2012.10.010. Epub 2012 Nov 2. Biochim Biophys Acta. 2013. PMID: 23128325 Review.
Cited by
-
Moonlighting nuclear pore proteins: tissue-specific nucleoporin function in health and disease.Histochem Cell Biol. 2018 Dec;150(6):593-605. doi: 10.1007/s00418-018-1748-8. Epub 2018 Oct 25. Histochem Cell Biol. 2018. PMID: 30361777 Review.
-
The Great Escape: mRNA Export through the Nuclear Pore Complex.Int J Mol Sci. 2021 Oct 29;22(21):11767. doi: 10.3390/ijms222111767. Int J Mol Sci. 2021. PMID: 34769195 Free PMC article. Review.
-
Genetic analysis of the Warburg effect in yeast.Adv Biol Regul. 2015 Jan;57:185-92. doi: 10.1016/j.jbior.2014.09.009. Epub 2014 Sep 30. Adv Biol Regul. 2015. PMID: 25446884 Free PMC article. Review.
-
Models of Distal Arthrogryposis and Lethal Congenital Contracture Syndrome.Genes (Basel). 2021 Jun 20;12(6):943. doi: 10.3390/genes12060943. Genes (Basel). 2021. PMID: 34203046 Free PMC article. Review.
-
Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform.Cell. 2016 Nov 17;167(5):1215-1228.e25. doi: 10.1016/j.cell.2016.10.028. Epub 2016 Nov 10. Cell. 2016. PMID: 27839866 Free PMC article.
References
-
- Al-Qattan MM, Shamseldin HE, Alkuraya FS. Familial dorsalization of the skin of the proximal palm and the instep of the sole of the foot. Gene. 2012;500:216–9. - PubMed
-
- Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nature Cell Biology. 2006;8:711–6. - PubMed
-
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–9. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

