GCN2-dependent metabolic stress is essential for endotoxemic cytokine induction and pathology
- PMID: 24248597
- PMCID: PMC3911502
- DOI: 10.1128/MCB.00946-13
GCN2-dependent metabolic stress is essential for endotoxemic cytokine induction and pathology
Abstract
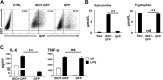
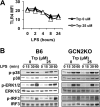
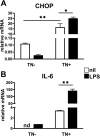
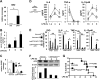
Activated inflammatory macrophages can express indoleamine 2,3-dioxygenase (IDO) and thus actively deplete their own tryptophan supply; however, it is not clear how amino acid depletion influences macrophage behavior in inflammatory environments. In this report, we demonstrate that the stress response kinase GCN2 promotes macrophage inflammation and mortality in a mouse model of septicemia. In vitro, enzymatic amino acid consumption enhanced sensitivity of macrophages to the Toll-like receptor 4 (TLR4) ligand lipopolysaccharide (LPS) with significantly increased interleukin 6 (IL-6) production. Tryptophan withdrawal induced the stress response proteins ATF4 and CHOP/GADD153; however, LPS stimulation rapidly enhanced expression of both proteins. Moreover, LPS-driven cytokine production under amino acid-deficient conditions was dependent on GCN2, as GCN2 knockout (GCN2KO) macrophages had a significant reduction of cytokine gene expression after LPS stimulation. To test the in vivo relevance of these findings, monocytic-lineage-specific GCN2KO mice were challenged with a lethal dose of LPS intraperitoneally (i.p.). The GCN2KO mice showed reduced inflammatory responses, with decreased IL-6 and IL-12 expression correlating with significant reduction in animal mortality. Thus, the data show that amino acid depletion stress signals (via GCN2) synergize with proinflammatory signals to potently increase innate immune responsiveness.
Figures




Similar articles
-
The amino acid sensor GCN2 inhibits inflammatory responses to apoptotic cells promoting tolerance and suppressing systemic autoimmunity.Proc Natl Acad Sci U S A. 2015 Aug 25;112(34):10774-9. doi: 10.1073/pnas.1504276112. Epub 2015 Aug 10. Proc Natl Acad Sci U S A. 2015. PMID: 26261340 Free PMC article.
-
Toll-Like Receptor 4 (TLR4) expression and stimulation in a model of intervertebral disc inflammation and degeneration.Spine (Phila Pa 1976). 2013 Jul 15;38(16):1343-51. doi: 10.1097/BRS.0b013e31826b71f4. Spine (Phila Pa 1976). 2013. PMID: 22850250
-
GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase.Immunity. 2005 May;22(5):633-42. doi: 10.1016/j.immuni.2005.03.013. Immunity. 2005. PMID: 15894280
-
Multiple Roles of the Stress Sensor GCN2 in Immune Cells.Int J Mol Sci. 2023 Feb 21;24(5):4285. doi: 10.3390/ijms24054285. Int J Mol Sci. 2023. PMID: 36901714 Free PMC article. Review.
-
Amino acid auxotrophy as a system of immunological control nodes.Nat Immunol. 2016 Feb;17(2):132-9. doi: 10.1038/ni.3323. Nat Immunol. 2016. PMID: 26784254 Free PMC article. Review.
Cited by
-
Dietary Intervention Impacts Immune Cell Functions and Dynamics by Inducing Metabolic Rewiring.Front Immunol. 2021 Feb 4;11:623989. doi: 10.3389/fimmu.2020.623989. eCollection 2020. Front Immunol. 2021. PMID: 33613560 Free PMC article. Review.
-
Meta-analysis of blood genome-wide expression profiling studies in pulmonary arterial hypertension.Am J Physiol Lung Cell Mol Physiol. 2020 Jan 1;318(1):L98-L111. doi: 10.1152/ajplung.00252.2019. Epub 2019 Oct 16. Am J Physiol Lung Cell Mol Physiol. 2020. PMID: 31617731 Free PMC article.
-
The amino acid sensor GCN2 inhibits inflammatory responses to apoptotic cells promoting tolerance and suppressing systemic autoimmunity.Proc Natl Acad Sci U S A. 2015 Aug 25;112(34):10774-9. doi: 10.1073/pnas.1504276112. Epub 2015 Aug 10. Proc Natl Acad Sci U S A. 2015. PMID: 26261340 Free PMC article.
-
New Insights into IDO Biology in Bacterial and Viral Infections.Front Immunol. 2014 Aug 11;5:384. doi: 10.3389/fimmu.2014.00384. eCollection 2014. Front Immunol. 2014. PMID: 25157255 Free PMC article. Review.
-
Bridging the divide: unveiling mutual immunological pathways of cancer and pregnancy.Inflamm Res. 2024 May;73(5):793-807. doi: 10.1007/s00011-024-01866-9. Epub 2024 Mar 16. Inflamm Res. 2024. PMID: 38492049 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
