Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes
- PMID: 24204277
- PMCID: PMC3814426
- DOI: 10.1371/journal.ppat.1003738
Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes
Abstract
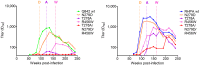
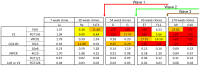
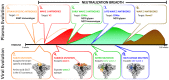
Identifying the targets of broadly neutralizing antibodies to HIV-1 and understanding how these antibodies develop remain important goals in the quest to rationally develop an HIV-1 vaccine. We previously identified a participant in the CAPRISA Acute Infection Cohort (CAP257) whose plasma neutralized 84% of heterologous viruses. In this study we showed that breadth in CAP257 was largely due to the sequential, transient appearance of three distinct broadly neutralizing antibody specificities spanning the first 4.5 years of infection. The first specificity targeted an epitope in the V2 region of gp120 that was also recognized by strain-specific antibodies 7 weeks earlier. Specificity for the autologous virus was determined largely by a rare N167 antigenic variant of V2, with viral escape to the more common D167 immunotype coinciding with the development of the first wave of broadly neutralizing antibodies. Escape from these broadly neutralizing V2 antibodies through deletion of the glycan at N160 was associated with exposure of an epitope in the CD4 binding site that became the target for a second wave of broadly neutralizing antibodies. Neutralization by these CD4 binding site antibodies was almost entirely dependent on the glycan at position N276. Early viral escape mutations in the CD4 binding site drove an increase in wave two neutralization breadth, as this second wave of heterologous neutralization matured to recognize multiple immunotypes within this site. The third wave targeted a quaternary epitope that did not overlap any of the four known sites of vulnerability on the HIV-1 envelope and remains undefined. Altogether this study showed that the human immune system is capable of generating multiple broadly neutralizing antibodies in response to a constantly evolving viral population that exposes new targets as a consequence of escape from earlier neutralizing antibodies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


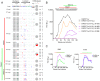

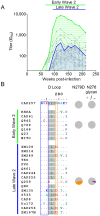
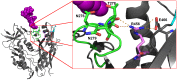
 denoting side chain contacts, and • denoting main chain and side chain contacts. Potential N-linked glycans are shaded grey. The frequency of escape mutations at position 279 (orange slices), in the N276/T278 glycosylation sequon (purple slices), or at position 456 (pink slices) for each time point is shown with pie charts.
denoting side chain contacts, and • denoting main chain and side chain contacts. Potential N-linked glycans are shaded grey. The frequency of escape mutations at position 279 (orange slices), in the N276/T278 glycosylation sequon (purple slices), or at position 456 (pink slices) for each time point is shown with pie charts.

Similar articles
-
Structure of an N276-Dependent HIV-1 Neutralizing Antibody Targeting a Rare V5 Glycan Hole Adjacent to the CD4 Binding Site.J Virol. 2016 Oct 28;90(22):10220-10235. doi: 10.1128/JVI.01357-16. Print 2016 Nov 15. J Virol. 2016. PMID: 27581986 Free PMC article.
-
Conformational Epitope-Specific Broadly Neutralizing Plasma Antibodies Obtained from an HIV-1 Clade C-Infected Elite Neutralizer Mediate Autologous Virus Escape through Mutations in the V1 Loop.J Virol. 2016 Jan 13;90(7):3446-57. doi: 10.1128/JVI.03090-15. J Virol. 2016. PMID: 26763999 Free PMC article.
-
Structure and Recognition of a Novel HIV-1 gp120-gp41 Interface Antibody that Caused MPER Exposure through Viral Escape.PLoS Pathog. 2017 Jan 11;13(1):e1006074. doi: 10.1371/journal.ppat.1006074. eCollection 2017 Jan. PLoS Pathog. 2017. PMID: 28076415 Free PMC article.
-
Structural Features of Broadly Neutralizing Antibodies and Rational Design of Vaccine.Adv Exp Med Biol. 2018;1075:73-95. doi: 10.1007/978-981-13-0484-2_4. Adv Exp Med Biol. 2018. PMID: 30030790 Review.
-
Survivors Remorse: antibody-mediated protection against HIV-1.Immunol Rev. 2017 Jan;275(1):271-284. doi: 10.1111/imr.12510. Immunol Rev. 2017. PMID: 28133809 Free PMC article. Review.
Cited by
-
Glycan-Dependent Neutralizing Antibodies Are Frequently Elicited in Individuals Chronically Infected with HIV-1 Clade B or C.AIDS Res Hum Retroviruses. 2015 Nov;31(11):1192-201. doi: 10.1089/AID.2015.0135. Epub 2015 Aug 5. AIDS Res Hum Retroviruses. 2015. PMID: 26149894 Free PMC article.
-
Shaping Polyclonal Responses via Antigen-Mediated Antibody Interference.iScience. 2020 Sep 17;23(10):101568. doi: 10.1016/j.isci.2020.101568. eCollection 2020 Oct 23. iScience. 2020. PMID: 33083735 Free PMC article.
-
Broadly neutralizing plasma antibodies effective against autologous circulating viruses in infants with multivariant HIV-1 infection.Nat Commun. 2020 Sep 2;11(1):4409. doi: 10.1038/s41467-020-18225-x. Nat Commun. 2020. PMID: 32879304 Free PMC article.
-
Common evolutionary features of the envelope glycoprotein of HIV-1 in patients belonging to a transmission chain.Sci Rep. 2020 Oct 7;10(1):16744. doi: 10.1038/s41598-020-73975-4. Sci Rep. 2020. PMID: 33028961 Free PMC article.
-
Association of mutations in V3/C3 domain with enhanced sensitivity of HIV-1 clade C primary envelopes to autologous broadly neutralizing plasma antibodies.Retrovirology. 2016 Jun 15;13(1):41. doi: 10.1186/s12977-016-0273-x. Retrovirology. 2016. PMID: 27307004 Free PMC article.
References
-
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, et al. (2003) Antibody neutralization and escape by HIV-1. Nature 422: 307–312. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

