A genomics-based classification of human lung tumors
- PMID: 24174329
- PMCID: PMC4006630
- DOI: 10.1126/scitranslmed.3006802
A genomics-based classification of human lung tumors
Abstract
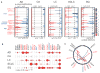
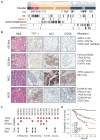
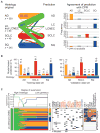
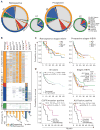
We characterized genome alterations in 1255 clinically annotated lung tumors of all histological subgroups to identify genetically defined and clinically relevant subtypes. More than 55% of all cases had at least one oncogenic genome alteration potentially amenable to specific therapeutic intervention, including several personalized treatment approaches that are already in clinical evaluation. Marked differences in the pattern of genomic alterations existed between and within histological subtypes, thus challenging the original histomorphological diagnosis. Immunohistochemical studies confirmed many of these reassigned subtypes. The reassignment eliminated almost all cases of large cell carcinomas, some of which had therapeutically relevant alterations. Prospective testing of our genomics-based diagnostic algorithm in 5145 lung cancer patients enabled a genome-based diagnosis in 3863 (75%) patients, confirmed the feasibility of rational reassignments of large cell lung cancer, and led to improvement in overall survival in patients with EGFR-mutant or ALK-rearranged cancers. Thus, our findings provide support for broad implementation of genome-based diagnosis of lung cancer.
Conflict of interest statement
Figures

Comment in
-
The evolution of tumor classification: a role for genomics?Cancer Cell. 2013 Dec 9;24(6):693-4. doi: 10.1016/j.ccr.2013.11.019. Cancer Cell. 2013. PMID: 24332038 Free PMC article.
Similar articles
-
Broad, Hybrid Capture-Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches.Clin Cancer Res. 2015 Aug 15;21(16):3631-9. doi: 10.1158/1078-0432.CCR-14-2683. Epub 2015 Jan 7. Clin Cancer Res. 2015. PMID: 25567908 Free PMC article.
-
Cancer genomics of lung cancer including malignant mesothelioma: A brief overview of current status and future prospects.Adv Biol Regul. 2020 Dec;78:100723. doi: 10.1016/j.jbior.2020.100723. Epub 2020 May 5. Adv Biol Regul. 2020. PMID: 32992231 Review.
-
Genomics AIDS lung tumor classification and treatment.Cancer Discov. 2014 Jan;4(1):OF5. doi: 10.1158/2159-8290.CD-NB2013-166. Epub 2013 Nov 21. Cancer Discov. 2014. PMID: 24402957
-
[Relevance of molecular alterations in histopathologic subtyping of lung adenocarcinoma based on 2011 International Multidisciplinary Lung Adenocarcinoma Classification].Zhonghua Bing Li Xue Za Zhi. 2012 Aug;41(8):505-10. doi: 10.3760/cma.j.issn.0529-5807.2012.08.001. Zhonghua Bing Li Xue Za Zhi. 2012. PMID: 23157740 Chinese.
-
Chromosomal and genomic changes in lung cancer.Cell Adh Migr. 2010 Jan-Mar;4(1):100-6. doi: 10.4161/cam.4.1.10884. Epub 2010 Jan 7. Cell Adh Migr. 2010. PMID: 20139701 Free PMC article. Review.
Cited by
-
Gene Amplification-Associated Overexpression of the Selenoprotein tRNA Enzyme TRIT1 Confers Sensitivity to Arsenic Trioxide in Small-Cell Lung Cancer.Cancers (Basel). 2021 Apr 14;13(8):1869. doi: 10.3390/cancers13081869. Cancers (Basel). 2021. PMID: 33919717 Free PMC article.
-
A literature review of a meta-analysis of BRAF mutations in non-small cell lung cancer.Medicine (Baltimore). 2024 Feb 23;103(8):e34654. doi: 10.1097/MD.0000000000034654. Medicine (Baltimore). 2024. PMID: 38394545 Free PMC article. Review.
-
ROS1 rearrangements in lung adenocarcinoma: prognostic impact, therapeutic options and genetic variability.Oncotarget. 2015 Apr 30;6(12):10577-85. doi: 10.18632/oncotarget.3387. Oncotarget. 2015. PMID: 25868855 Free PMC article.
-
The clinical benefit of molecular re-assessments in management of progressive lung cancer.Transl Lung Cancer Res. 2021 Mar;10(3):1582-1587. doi: 10.21037/tlcr-20-996. Transl Lung Cancer Res. 2021. PMID: 33889532 Free PMC article.
-
NRF2 Mediates Cellular Resistance to Transformation, Radiation, and Inflammation in Mice.Antioxidants (Basel). 2022 Aug 25;11(9):1649. doi: 10.3390/antiox11091649. Antioxidants (Basel). 2022. PMID: 36139722 Free PMC article.
References
-
- Travis W, Brambilla E, Mueller-Hermelink H, Harris C. World Health Organization Classification of Tumours. Pathology and Genetics: Tumours of the Lung, Pleura, Thymus and Heart. World Health Organization; Geneva: 2004.
-
- Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger K, Yatabe Y, Powell CA, Beer D, Riely G, Garg K, Austin JH, Rusch VW, Hirsch FR, Jett J, Yang PC, Gould M. American Thoracic Society, International Association for the Study of Lung Cancer/ American Thoracic Society/European Respiratory Society: International multidisciplinary classification of lung adenocarcinoma: Executive summary. Proc Am Thorac Soc. 2011;8:381–385. - PubMed
-
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. - PubMed
-
- Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. - PubMed
-
- Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

