Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study
- PMID: 24167453
- PMCID: PMC3805485
- DOI: 10.1371/journal.pmed.1001538
Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study
Abstract
Background: A major impediment to tuberculosis control in Africa is the difficulty in diagnosing active tuberculosis (TB), particularly in the context of HIV infection. We hypothesized that a unique host blood RNA transcriptional signature would distinguish TB from other diseases (OD) in HIV-infected and -uninfected patients, and that this could be the basis of a simple diagnostic test.
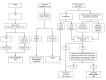
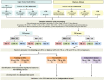
Methods and findings: Adult case-control cohorts were established in South Africa and Malawi of HIV-infected or -uninfected individuals consisting of 584 patients with either TB (confirmed by culture of Mycobacterium tuberculosis [M.TB] from sputum or tissue sample in a patient under investigation for TB), OD (i.e., TB was considered in the differential diagnosis but then excluded), or healthy individuals with latent TB infection (LTBI). Individuals were randomized into training (80%) and test (20%) cohorts. Blood transcriptional profiles were assessed and minimal sets of significantly differentially expressed transcripts distinguishing TB from LTBI and OD were identified in the training cohort. A 27 transcript signature distinguished TB from LTBI and a 44 transcript signature distinguished TB from OD. To evaluate our signatures, we used a novel computational method to calculate a disease risk score (DRS) for each patient. The classification based on this score was first evaluated in the test cohort, and then validated in an independent publically available dataset (GSE19491). In our test cohort, the DRS classified TB from LTBI (sensitivity 95%, 95% CI [87-100]; specificity 90%, 95% CI [80-97]) and TB from OD (sensitivity 93%, 95% CI [83-100]; specificity 88%, 95% CI [74-97]). In the independent validation cohort, TB patients were distinguished both from LTBI individuals (sensitivity 95%, 95% CI [85-100]; specificity 94%, 95% CI [84-100]) and OD patients (sensitivity 100%, 95% CI [100-100]; specificity 96%, 95% CI [93-100]). Limitations of our study include the use of only culture confirmed TB patients, and the potential that TB may have been misdiagnosed in a small proportion of OD patients despite the extensive clinical investigation used to assign each patient to their diagnostic group.
Conclusions: In our study, blood transcriptional signatures distinguished TB from other conditions prevalent in HIV-infected and -uninfected African adults. Our DRS, based on these signatures, could be developed as a test for TB suitable for use in HIV endemic countries. Further evaluation of the performance of the signatures and DRS in prospective populations of patients with symptoms consistent with TB will be needed to define their clinical value under operational conditions. Please see later in the article for the Editors' Summary.
Conflict of interest statement
The authors have declared that patent applications have been filed for the Disease Risk score (GB1201766.1) and TB/LTBI and TB/OD signatures (GB1213636.2).
Figures



Comment in
-
A transcriptional signature for active TB: have we found the needle in the haystack?PLoS Med. 2013 Oct;10(10):e1001539. doi: 10.1371/journal.pmed.1001539. Epub 2013 Oct 22. PLoS Med. 2013. PMID: 24167454 Free PMC article.
Similar articles
-
Diagnosis of childhood tuberculosis and host RNA expression in Africa.N Engl J Med. 2014 May 1;370(18):1712-1723. doi: 10.1056/NEJMoa1303657. N Engl J Med. 2014. PMID: 24785206 Free PMC article.
-
Assessment of Validity of a Blood-Based 3-Gene Signature Score for Progression and Diagnosis of Tuberculosis, Disease Severity, and Treatment Response.JAMA Netw Open. 2018 Oct 5;1(6):e183779. doi: 10.1001/jamanetworkopen.2018.3779. JAMA Netw Open. 2018. PMID: 30646264 Free PMC article.
-
CCL1 and IL-2Ra differentiate Tuberculosis disease from latent infection Irrespective of HIV infection in low TB burden countries.J Infect. 2021 Oct;83(4):433-443. doi: 10.1016/j.jinf.2021.07.036. Epub 2021 Jul 29. J Infect. 2021. PMID: 34333033
-
[Evolution of IGRA researches].Kekkaku. 2008 Sep;83(9):641-52. Kekkaku. 2008. PMID: 18979999 Review. Japanese.
-
[Characteristics of a diagnostic method for tuberculosis infection based on whole blood interferon-gamma assay].Kekkaku. 2006 Nov;81(11):681-6. Kekkaku. 2006. PMID: 17154047 Review. Japanese.
Cited by
-
A blood-based host gene expression assay for early detection of respiratory viral infection: an index-cluster prospective cohort study.Lancet Infect Dis. 2021 Mar;21(3):396-404. doi: 10.1016/S1473-3099(20)30486-2. Epub 2020 Sep 24. Lancet Infect Dis. 2021. PMID: 32979932 Free PMC article.
-
A qPCR expression assay of IFI44L gene differentiates viral from bacterial infections in febrile children.Sci Rep. 2019 Aug 13;9(1):11780. doi: 10.1038/s41598-019-48162-9. Sci Rep. 2019. PMID: 31409879 Free PMC article.
-
Incipient and Subclinical Tuberculosis: a Clinical Review of Early Stages and Progression of Infection.Clin Microbiol Rev. 2018 Jul 18;31(4):e00021-18. doi: 10.1128/CMR.00021-18. Print 2018 Oct. Clin Microbiol Rev. 2018. PMID: 30021818 Free PMC article. Review.
-
Monocyte-to-Lymphocyte Ratio Is Associated With Tuberculosis Disease and Declines With Anti-TB Treatment in HIV-Infected Children.J Acquir Immune Defic Syndr. 2019 Feb 1;80(2):174-181. doi: 10.1097/QAI.0000000000001893. J Acquir Immune Defic Syndr. 2019. PMID: 30399036 Free PMC article. Clinical Trial.
-
Personalised Medicine for Tuberculosis and Non-Tuberculous Mycobacterial Pulmonary Disease.Microorganisms. 2021 Oct 26;9(11):2220. doi: 10.3390/microorganisms9112220. Microorganisms. 2021. PMID: 34835346 Free PMC article. Review.
References
-
- Munthali L, Mwaungulu JN, Munthali K, Bowie C, Crampin AC (2006) Using tuberculosis suspects to identify patients eligible for antiretroviral treatment. Int J Tuberc Lung Dis 10: 199–202. - PubMed
-
- Aabye MG, Ravn P, PrayGod G, Jeremiah K, Mugomela A, et al. (2009) The impact of HIV infection and CD4 cell count on the performance of an interferon gamma release assay in patients with pulmonary tuberculosis. PLoS One 4: e4220 doi:10.1371/journal.pone.0004220 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

