The molecular recognition of kink-turn structure by the L7Ae class of proteins
- PMID: 24149842
- PMCID: PMC3884654
- DOI: 10.1261/rna.041517.113
The molecular recognition of kink-turn structure by the L7Ae class of proteins
Abstract
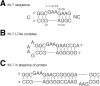
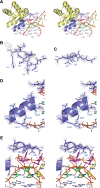
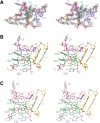
L7Ae is a member of a protein family that binds kink-turns (k-turns) in many functional RNA species. We have solved the X-ray crystal structure of the near-consensus sequence Kt-7 of Haloarcula marismortui bound by Archaeoglobus fulgidus L7Ae at 2.3-Å resolution. We also present a structure of Kt-7 in the absence of bound protein at 2.2-Å resolution. As a result, we can describe a general mode of recognition of k-turn structure by the L7Ae family proteins. The protein makes interactions in the widened major groove on the outer face of the k-turn. Two regions of the protein are involved. One is an α-helix that enters the major groove of the NC helix, making both nonspecific backbone interactions and specific interactions with the guanine nucleobases of the conserved G • A pairs. A hydrophobic loop makes close contact with the L1 and L2 bases, and a glutamate side chain hydrogen bonds with L1. Taken together, these interactions are highly selective for the structure of the k-turn and suggest how conformational selection of the folded k-turn occurs.
Keywords: RNA structure; RNA-protein recognition; X-ray crystallography; k-turn motif.
Figures
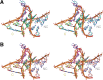

Similar articles
-
The role of RNA structure in translational regulation by L7Ae protein in archaea.RNA. 2019 Jan;25(1):60-69. doi: 10.1261/rna.068510.118. Epub 2018 Oct 16. RNA. 2019. PMID: 30327333 Free PMC article.
-
The archaeal sRNA binding protein L7Ae has a 3D structure very similar to that of its eukaryal counterpart while having a broader RNA-binding specificity.J Mol Biol. 2004 Sep 17;342(3):757-73. doi: 10.1016/j.jmb.2004.07.046. J Mol Biol. 2004. PMID: 15342235
-
The importance of G.A hydrogen bonding in the metal ion- and protein-induced folding of a kink turn RNA.J Mol Biol. 2008 Aug 29;381(2):431-42. doi: 10.1016/j.jmb.2008.05.052. Epub 2008 May 29. J Mol Biol. 2008. PMID: 18603260
-
The kink-turn in the structural biology of RNA.Q Rev Biophys. 2018 Jan;51:e5. doi: 10.1017/S0033583518000033. Q Rev Biophys. 2018. PMID: 30912490 Review.
-
The Kink Turn, a Key Architectural Element in RNA Structure.J Mol Biol. 2016 Feb 27;428(5 Pt A):790-801. doi: 10.1016/j.jmb.2015.09.026. Epub 2015 Oct 29. J Mol Biol. 2016. PMID: 26522935 Free PMC article. Review.
Cited by
-
Simulation Study of the Plasticity of k-Turn Motif in Different Environments.Biophys J. 2020 Oct 6;119(7):1416-1426. doi: 10.1016/j.bpj.2020.08.015. Epub 2020 Aug 20. Biophys J. 2020. PMID: 32918889 Free PMC article.
-
Lokiarchaeota Marks the Transition between the Archaeal and Eukaryotic Selenocysteine Encoding Systems.Mol Biol Evol. 2016 Sep;33(9):2441-53. doi: 10.1093/molbev/msw122. Epub 2016 Jul 12. Mol Biol Evol. 2016. PMID: 27413050 Free PMC article.
-
Sequence Analysis and Comparative Study of the Protein Subunits of Archaeal RNase P.Biomolecules. 2016 Apr 20;6(2):22. doi: 10.3390/biom6020022. Biomolecules. 2016. PMID: 27104580 Free PMC article. Review.
-
A novel double kink-turn module in euryarchaeal RNase P RNAs.Nucleic Acids Res. 2017 Jul 7;45(12):7432-7440. doi: 10.1093/nar/gkx388. Nucleic Acids Res. 2017. PMID: 28525600 Free PMC article.
-
The role of RNA structure in translational regulation by L7Ae protein in archaea.RNA. 2019 Jan;25(1):60-69. doi: 10.1261/rna.068510.118. Epub 2018 Oct 16. RNA. 2019. PMID: 30327333 Free PMC article.
References
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. - PubMed
-
- Beaucage SL, Caruthers MH. 1981. Deoxynucleoside phosphoramidites—A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett 22: 1859–1862.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
