Highlights of the DNA cutters: a short history of the restriction enzymes
- PMID: 24141096
- PMCID: PMC3874209
- DOI: 10.1093/nar/gkt990
Highlights of the DNA cutters: a short history of the restriction enzymes
Abstract
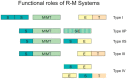
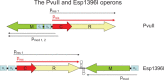
In the early 1950's, 'host-controlled variation in bacterial viruses' was reported as a non-hereditary phenomenon: one cycle of viral growth on certain bacterial hosts affected the ability of progeny virus to grow on other hosts by either restricting or enlarging their host range. Unlike mutation, this change was reversible, and one cycle of growth in the previous host returned the virus to its original form. These simple observations heralded the discovery of the endonuclease and methyltransferase activities of what are now termed Type I, II, III and IV DNA restriction-modification systems. The Type II restriction enzymes (e.g. EcoRI) gave rise to recombinant DNA technology that has transformed molecular biology and medicine. This review traces the discovery of restriction enzymes and their continuing impact on molecular biology and medicine.
Figures


Similar articles
-
Type III restriction-modification enzymes: a historical perspective.Nucleic Acids Res. 2014 Jan;42(1):45-55. doi: 10.1093/nar/gkt616. Epub 2013 Jul 17. Nucleic Acids Res. 2014. PMID: 23863841 Free PMC article. Review.
-
Two-step cloning and expression in Escherichia coli of the DNA restriction-modification system StyLTI of Salmonella typhimurium.J Bacteriol. 1991 Feb;173(3):1321-7. doi: 10.1128/jb.173.3.1321-1327.1991. J Bacteriol. 1991. PMID: 1846861 Free PMC article.
-
Complex restriction enzymes: NTP-driven molecular motors.Biochimie. 2002 Nov;84(11):1047-59. doi: 10.1016/s0300-9084(02)00020-2. Biochimie. 2002. PMID: 12595133 Review.
-
Restriction Enzymes.Cold Spring Harb Protoc. 2021 Apr 1;2021(4). doi: 10.1101/pdb.top101360. Cold Spring Harb Protoc. 2021. PMID: 33536287
-
Solitary restriction endonucleases in prokaryotic genomes.Nucleic Acids Res. 2012 Nov 1;40(20):10107-15. doi: 10.1093/nar/gks853. Epub 2012 Sep 10. Nucleic Acids Res. 2012. PMID: 22965118 Free PMC article.
Cited by
-
Impacts of Mycoplasma agalactiae restriction-modification systems on pan-epigenome dynamics and genome plasticity.Microb Genom. 2022 May;8(5):mgen000829. doi: 10.1099/mgen.0.000829. Microb Genom. 2022. PMID: 35576144 Free PMC article.
-
A model for the evolution of prokaryotic DNA restriction-modification systems based upon the structural malleability of Type I restriction-modification enzymes.Nucleic Acids Res. 2018 Sep 28;46(17):9067-9080. doi: 10.1093/nar/gky760. Nucleic Acids Res. 2018. PMID: 30165537 Free PMC article.
-
Barriers to genetic manipulation of Enterococci: Current Approaches and Future Directions.FEMS Microbiol Rev. 2022 Nov 2;46(6):fuac036. doi: 10.1093/femsre/fuac036. FEMS Microbiol Rev. 2022. PMID: 35883217 Free PMC article. Review.
-
Methylome evolution suggests lineage-dependent selection in the gastric pathogen Helicobacter pylori.Commun Biol. 2023 Aug 12;6(1):839. doi: 10.1038/s42003-023-05218-x. Commun Biol. 2023. PMID: 37573385 Free PMC article.
-
A simple and dual expression plasmid system in prokaryotic (E. coli) and mammalian cells.PLoS One. 2019 May 2;14(5):e0216169. doi: 10.1371/journal.pone.0216169. eCollection 2019. PLoS One. 2019. PMID: 31048860 Free PMC article.
References
-
- Roberts RJ, Cheng X. Base flipping. Annu. Rev. Biochem. 1998;67:181–198. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

