Bi-functional cross-linking reagents efficiently capture protein-DNA complexes in Drosophila embryos
- PMID: 24135698
- PMCID: PMC3974894
- DOI: 10.4161/fly.26805
Bi-functional cross-linking reagents efficiently capture protein-DNA complexes in Drosophila embryos
Abstract
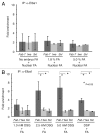
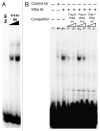
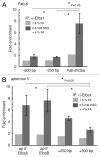
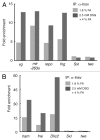
Chromatin immunoprecipitation (ChIP) is widely used for mapping DNA-protein interactions across eukaryotic genomes in cells, tissues or even whole organisms. Critical to this procedure is the efficient cross-linking of chromatin-associated proteins to DNA sequences that are in close proximity. Since the mid-nineties formaldehyde fixation has been the method of choice. However, some protein-DNA complexes cannot be successfully captured for ChIP using formaldehyde. One such formaldehyde refractory complex is the developmentally regulated insulator factor, Elba. Here we describe a new embryo fixation procedure using the bi-functional cross-linking reagents DSG (disuccinimidyl glutarate) and DSP (dithiobis[succinimidyl propionate). We show that unlike standard formaldehyde fixation protocols, it is possible to capture Elba association with insulator elements in 2-5 h embryos using this new cross-linking procedure. We show that this new cross-linking procedure can also be applied to localize nuclear proteins that are amenable to ChIP using standard formaldehyde cross-linking protocols, and that in the cases tested the enrichment was generally superior to that achieved using formaldehyde cross-linking.
Keywords: ChIP; DNA binding; DSG DSP; Elba; Insensitive; bi-functional cross-linkers; chromatin immunoprecipitation; formadelhyde; insulators.
Figures
Similar articles
-
Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation.Biotechniques. 2005 Nov;39(5):715-25. doi: 10.2144/000112014. Biotechniques. 2005. PMID: 16315372
-
ChIP bias as a function of cross-linking time.Chromosome Res. 2016 May;24(2):175-81. doi: 10.1007/s10577-015-9509-1. Epub 2015 Dec 21. Chromosome Res. 2016. PMID: 26685864 Free PMC article.
-
Cleavable Crosslinkers as Tissue Fixation Reagents for Proteomic Analysis.Chembiochem. 2018 Apr 4;19(7):736-743. doi: 10.1002/cbic.201700625. Epub 2018 Feb 16. Chembiochem. 2018. PMID: 29356267
-
Fish'n ChIPs: chromatin immunoprecipitation in the zebrafish embryo.Methods Mol Biol. 2009;567:75-86. doi: 10.1007/978-1-60327-414-2_5. Methods Mol Biol. 2009. PMID: 19588086 Review.
-
Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo.Curr Protoc Cell Biol. 2004 Sep;Chapter 17:Unit 17.7. doi: 10.1002/0471143030.cb1707s23. Curr Protoc Cell Biol. 2004. PMID: 18228445 Review.
Cited by
-
BEN-solo factors partition active chromatin to ensure proper gene activation in Drosophila.Nat Commun. 2019 Dec 13;10(1):5700. doi: 10.1038/s41467-019-13558-8. Nat Commun. 2019. PMID: 31836703 Free PMC article.
-
Photocaged Quinone Methide Crosslinkers for Light-Controlled Chemical Crosslinking of Protein-Protein and Protein-DNA Complexes.Angew Chem Int Ed Engl. 2019 Dec 19;58(52):18839-18843. doi: 10.1002/anie.201910135. Epub 2019 Nov 8. Angew Chem Int Ed Engl. 2019. PMID: 31644827 Free PMC article.
-
Optimized ChIP-seq method facilitates transcription factor profiling in human tumors.Life Sci Alliance. 2018 Dec 28;2(1):e201800115. doi: 10.26508/lsa.201800115. eCollection 2019 Feb. Life Sci Alliance. 2018. PMID: 30620009 Free PMC article.
-
The BEN Domain Protein Insensitive Binds to the Fab-7 Chromatin Boundary To Establish Proper Segmental Identity in Drosophila.Genetics. 2018 Oct;210(2):573-585. doi: 10.1534/genetics.118.301259. Epub 2018 Aug 6. Genetics. 2018. PMID: 30082280 Free PMC article.
-
Identification and partial characterization of new cell density-dependent nucleocytoplasmic shuttling proteins and open chromatin.Sci Rep. 2023 Dec 8;13(1):21723. doi: 10.1038/s41598-023-49100-6. Sci Rep. 2023. PMID: 38066085 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
