Use of principal components analysis and protein microarray to explore the association of HIV-1-specific IgG responses with disease progression
- PMID: 24134221
- PMCID: PMC3931433
- DOI: 10.1089/AID.2013.0088
Use of principal components analysis and protein microarray to explore the association of HIV-1-specific IgG responses with disease progression
Abstract
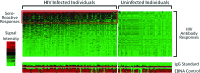
The role of HIV-1-specific antibody responses in HIV disease progression is complex and would benefit from analysis techniques that examine clusterings of responses. Protein microarray platforms facilitate the simultaneous evaluation of numerous protein-specific antibody responses, though excessive data are cumbersome in analyses. Principal components analysis (PCA) reduces data dimensionality by generating fewer composite variables that maximally account for variance in a dataset. To identify clusters of antibody responses involved in disease control, we investigated the association of HIV-1-specific antibody responses by protein microarray, and assessed their association with disease progression using PCA in a nested cohort design. Associations observed among collections of antibody responses paralleled protein-specific responses. At baseline, greater antibody responses to the transmembrane glycoprotein (TM) and reverse transcriptase (RT) were associated with higher viral loads, while responses to the surface glycoprotein (SU), capsid (CA), matrix (MA), and integrase (IN) proteins were associated with lower viral loads. Over 12 months greater antibody responses were associated with smaller decreases in CD4 count (CA, MA, IN), and reduced likelihood of disease progression (CA, IN). PCA and protein microarray analyses highlighted a collection of HIV-specific antibody responses that together were associated with reduced disease progression, and may not have been identified by examining individual antibody responses. This technique may be useful to explore multifaceted host-disease interactions, such as HIV coinfections.
Trial registration: ClinicalTrials.gov NCT00507221.
Figures


Similar articles
-
Pol-Driven Replicative Capacity Impacts Disease Progression in HIV-1 Subtype C Infection.J Virol. 2018 Sep 12;92(19):e00811-18. doi: 10.1128/JVI.00811-18. Print 2018 Oct 1. J Virol. 2018. PMID: 29997209 Free PMC article.
-
Frequencies of Circulating Th1-Biased T Follicular Helper Cells in Acute HIV-1 Infection Correlate with the Development of HIV-Specific Antibody Responses and Lower Set Point Viral Load.J Virol. 2018 Jul 17;92(15):e00659-18. doi: 10.1128/JVI.00659-18. Print 2018 Aug 1. J Virol. 2018. PMID: 29793949 Free PMC article.
-
Association between different anti-Tat antibody isotypes and HIV disease progression: data from an African cohort.BMC Infect Dis. 2016 Jul 22;16:344. doi: 10.1186/s12879-016-1647-3. BMC Infect Dis. 2016. PMID: 27450538 Free PMC article.
-
Antibodies against full-length Tat protein and some low-molecular-weight Tat-peptides correlate with low or undetectable viral load in HIV-1 seropositive patients.J Clin Virol. 2001 Apr;21(1):81-9. doi: 10.1016/s1386-6532(00)00189-x. J Clin Virol. 2001. PMID: 11255101
-
Optimal conditions of immune complex transfer enzyme immunoassays for antibody IgGs to HIV-1 using recombinant p17, p24, and reverse transcriptase as antigens.J Clin Lab Anal. 1998;12(2):98-107. doi: 10.1002/(SICI)1098-2825(1998)12:2<98::AID-JCLA5>3.0.CO;2-F. J Clin Lab Anal. 1998. PMID: 9524294 Free PMC article.
Cited by
-
A systems biology approach for diagnostic and vaccine antigen discovery in tropical infectious diseases.Curr Opin Infect Dis. 2015 Oct;28(5):438-45. doi: 10.1097/QCO.0000000000000193. Curr Opin Infect Dis. 2015. PMID: 26237545 Free PMC article. Review.
-
A targeted immunomic approach identifies diagnostic antigens in the human pathogen Babesia microti.Transfusion. 2016 Aug;56(8):2085-99. doi: 10.1111/trf.13640. Epub 2016 May 17. Transfusion. 2016. PMID: 27184823 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

