MEGA6: Molecular Evolutionary Genetics Analysis version 6.0
- PMID: 24132122
- PMCID: PMC3840312
- DOI: 10.1093/molbev/mst197
MEGA6: Molecular Evolutionary Genetics Analysis version 6.0
Abstract
We announce the release of an advanced version of the Molecular Evolutionary Genetics Analysis (MEGA) software, which currently contains facilities for building sequence alignments, inferring phylogenetic histories, and conducting molecular evolutionary analysis. In version 6.0, MEGA now enables the inference of timetrees, as it implements the RelTime method for estimating divergence times for all branching points in a phylogeny. A new Timetree Wizard in MEGA6 facilitates this timetree inference by providing a graphical user interface (GUI) to specify the phylogeny and calibration constraints step-by-step. This version also contains enhanced algorithms to search for the optimal trees under evolutionary criteria and implements a more advanced memory management that can double the size of sequence data sets to which MEGA can be applied. Both GUI and command-line versions of MEGA6 can be downloaded from www.megasoftware.net free of charge.
Keywords: phylogeny; relaxed clocks; software.
Figures
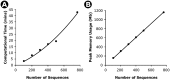

Similar articles
-
MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets.Mol Biol Evol. 2016 Jul;33(7):1870-4. doi: 10.1093/molbev/msw054. Epub 2016 Mar 22. Mol Biol Evol. 2016. PMID: 27004904 Free PMC article.
-
MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods.Mol Biol Evol. 2011 Oct;28(10):2731-9. doi: 10.1093/molbev/msr121. Epub 2011 May 4. Mol Biol Evol. 2011. PMID: 21546353 Free PMC article.
-
MEGA11: Molecular Evolutionary Genetics Analysis Version 11.Mol Biol Evol. 2021 Jun 25;38(7):3022-3027. doi: 10.1093/molbev/msab120. Mol Biol Evol. 2021. PMID: 33892491 Free PMC article.
-
MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0.Mol Biol Evol. 2007 Aug;24(8):1596-9. doi: 10.1093/molbev/msm092. Epub 2007 May 7. Mol Biol Evol. 2007. PMID: 17488738
-
MEGA biocentric software for sequence and phylogenetic analysis: a review.Int J Bioinform Res Appl. 2010;6(3):230-40. doi: 10.1504/IJBRA.2010.034072. Int J Bioinform Res Appl. 2010. PMID: 20615832 Review.
Cited by
-
Analyzes of pan-genome and resequencing atlas unveil the genetic basis of jujube domestication.Nat Commun. 2024 Oct 29;15(1):9320. doi: 10.1038/s41467-024-53718-z. Nat Commun. 2024. PMID: 39472552 Free PMC article.
-
First detection of D181 genotype of infectious bronchitis in poultry flocks of Morocco.Virol J. 2024 Oct 28;21(1):267. doi: 10.1186/s12985-024-02539-z. Virol J. 2024. PMID: 39468571 Free PMC article.
-
Molecular cloning and biochemical characterization of indole-3-acetic acid methyltransferase from Japanese star anise (Illicium anisatum).Plant Biotechnol (Tokyo). 2024 Mar 25;41(1):65-70. doi: 10.5511/plantbiotechnology.23.1224a. Plant Biotechnol (Tokyo). 2024. PMID: 39464862 Free PMC article.
-
Insights into Genes Encoding LEA_1 Domain-Containing Proteins in Cyperus esculentus, a Desiccation-Tolerant Tuber Plant.Plants (Basel). 2024 Oct 19;13(20):2933. doi: 10.3390/plants13202933. Plants (Basel). 2024. PMID: 39458880 Free PMC article.
-
FgUbiH Is Essential for Vegetative Development, Energy Metabolism, and Antioxidant Activity in Fusarium graminearum.Microorganisms. 2024 Oct 20;12(10):2093. doi: 10.3390/microorganisms12102093. Microorganisms. 2024. PMID: 39458403 Free PMC article.
References
-
- Hedges SB, Kumar S. Precision of molecular time estimates. Trends Genet. 2004;20:242–247. - PubMed
-
- Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput Appl Biosci. 1994;10:189–191. - PubMed
-
- Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford; New York: Oxford University Press; 2000.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

