Molecular cloning and characterization of SL3: a stem cell-specific SL RNA from the planarian Schmidtea mediterranea
- PMID: 24120894
- PMCID: PMC3994195
- DOI: 10.1016/j.gene.2013.09.101
Molecular cloning and characterization of SL3: a stem cell-specific SL RNA from the planarian Schmidtea mediterranea
Abstract
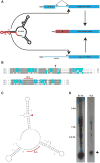
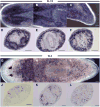
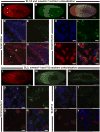
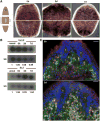
Spliced leader (SL) trans-splicing is a biological phenomenon, common among many metazoan taxa, consisting in the transfer of a short leader sequence from a small SL RNA to the 5' end of a subset of pre-mRNAs. While knowledge of the biochemical mechanisms driving this process has accumulated over the years, the functional consequences of such post-transcriptional event at the organismal level remain unclear. In addition, the fact that functional analyses have been undertaken mainly in trypanosomes and nematodes leaves a somehow fragmented picture of the possible biological significance and evolution of SL trans-splicing in eukaryotes. Here, we analyzed the spatial expression of SL RNAs in the planarian flatworm Schmidtea mediterranea, with the goal of identifying novel developmental paradigms for the study of trans-splicing in metazoans. Besides the previously identified SL1 and SL2, S. mediterranea expresses a third SL RNA described here as SL3. While, SL1 and SL2 are collectively expressed in a broad range of planarian cell types, SL3 is highly enriched in a subset of the planarian stem cells engaged in regenerative responses. Our findings provide new opportunities to study how trans-splicing may regulate the phenotype of a cell.
Keywords: 0.15M NaCl/0.015M Na(3)·citrate pH7.6; 2,2,7-trimethylguanosine; 4′,6-diamidino-2-phenylindole; 5-bromo-4-chloro-3-indolyl phosphate; 7-methylguanosine; A; ATP; BAC; BCIP; C; CB; DAPI; DIG; DNA complementary to RNA; DNP; EST; FISH; FITC; G; IRR; Kb; MMG; NBT; Nitro Blue tetrazolium; OCT; Optimal Cutting Temperature compound; PBS; PCR; Planarians; R; RACE; RNP; RT-PCR; Regeneration; S; SDS; SL; SL RNA; SL RNP; SSC; Schmidtea mediterranea piwi-like protein; Sm; Smith's antigen; Stem cells; T; TMG; Trans-splicing; U; UTR; WT; adenosine; adenosine triphosphate; b; bacterial artificial chromosome; base pair(s); bp; brain; cDNA; cNeoblast; chromatoid bodies; clonogenic neoblast; cytidine; d; deoxyribo; digoxigenin; dinitrophenol; expressed sequence tag; fluorescein isothiocyanate; fluorescent in situ hybridization; g; guanosine; gut; in vitro transcribed standard; irradiation; kilobase(s) or 1000bp; mRNA; messenger RNA; nc; nerve chords; nt; nucleotide(s); p; pharynx; phosphate buffered saline; photoreceptors; polymerase chain reaction; pr; purine; rNTP; rRNA; rapid amplification of cDNA ends; reverse transcriptase-PCR; ribonucleoprotein; ribonucleotide; ribosomal RNA; sDMA; small nuclear RNA; smedwi-1; snRNA; sodium dodecyl sulfate; spliced leader; spliced leader RNA; spliced leader RNA ribonucleoprotein; symmetrical dimethylarginine; thymidine; untranslated region(s); uridine; wild type.
© 2013 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Surprising diversity and distribution of spliced leader RNAs in flatworms.Mol Biochem Parasitol. 1997 Jul;87(1):29-48. doi: 10.1016/s0166-6851(97)00040-6. Mol Biochem Parasitol. 1997. PMID: 9233671
-
Spliced leader RNA-mediated trans-splicing in phylum Rotifera.Mol Biol Evol. 2005 Jun;22(6):1482-9. doi: 10.1093/molbev/msi139. Epub 2005 Mar 23. Mol Biol Evol. 2005. PMID: 15788744
-
Spliced-leader trans-splicing in freshwater planarians.Mol Biol Evol. 2005 Oct;22(10):2048-54. doi: 10.1093/molbev/msi200. Epub 2005 Jun 22. Mol Biol Evol. 2005. PMID: 15972844
-
On the Possibility of an Early Evolutionary Origin for the Spliced Leader Trans-Splicing.J Mol Evol. 2017 Aug;85(1-2):37-45. doi: 10.1007/s00239-017-9803-y. Epub 2017 Jul 25. J Mol Evol. 2017. PMID: 28744787 Review.
-
Planarian flatworms as a new model system for understanding the epigenetic regulation of stem cell pluripotency and differentiation.Semin Cell Dev Biol. 2019 Mar;87:79-94. doi: 10.1016/j.semcdb.2018.04.007. Epub 2018 Apr 27. Semin Cell Dev Biol. 2019. PMID: 29694837 Review.
Cited by
-
Heterologous reporter expression in the planarian Schmidtea mediterranea through somatic mRNA transfection.Cell Rep Methods. 2022 Sep 20;2(10):100298. doi: 10.1016/j.crmeth.2022.100298. eCollection 2022 Oct 24. Cell Rep Methods. 2022. PMID: 36313809 Free PMC article.
-
A diversified and segregated mRNA spliced-leader system in the parasitic Perkinsozoa.Open Biol. 2022 Aug;12(8):220126. doi: 10.1098/rsob.220126. Epub 2022 Aug 24. Open Biol. 2022. PMID: 36000319 Free PMC article.
-
Planarian stem cells specify fate yet retain potency during the cell cycle.Cell Stem Cell. 2021 Jul 1;28(7):1307-1322.e5. doi: 10.1016/j.stem.2021.03.021. Epub 2021 Apr 20. Cell Stem Cell. 2021. PMID: 33882291 Free PMC article.
-
Complete representation of a tapeworm genome reveals chromosomes capped by centromeres, necessitating a dual role in segregation and protection.BMC Biol. 2020 Nov 9;18(1):165. doi: 10.1186/s12915-020-00899-w. BMC Biol. 2020. PMID: 33167983 Free PMC article.
-
SLFinder, a pipeline for the novel identification of splice-leader sequences: a good enough solution for a complex problem.BMC Bioinformatics. 2020 Jul 8;21(1):293. doi: 10.1186/s12859-020-03610-6. BMC Bioinformatics. 2020. PMID: 32640978 Free PMC article.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Blumenthal T. Trans-splicing and operons. WormBook: the online review of C elegans biology. 2005:1–9. - PubMed
-
- Boothroyd JC, Cross GA. Transcripts coding for variant surface glycoproteins of Trypanosoma brucei have a short, identical exon at their 5′ end. Gene. 1982;20:281–289. - PubMed
-
- Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Luhrmann R. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem. 2000;275:17122–17129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

