Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling
- PMID: 24119841
- PMCID: PMC3881181
- DOI: 10.1016/j.cell.2013.09.049
Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling
Abstract
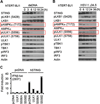
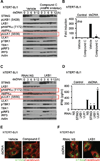
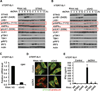
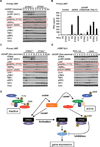
Activation of the stimulator of interferon genes (STING) pathway by microbial or self-DNA, as well as cyclic dinucleotides (CDNs), results in the induction of numerous genes that suppress pathogen replication and facilitate adaptive immunity. However, sustained gene transcription is rigidly prevented to avoid lethal STING-dependent proinflammatory disease by mechanisms that remain unknown. We demonstrate here that, after autophagy-dependent STING delivery of TANK-binding kinase 1 (TBK1) to endosomal/lysosomal compartments and activation of transcription factors interferon regulatory factor 3 (IRF3) and NF-κB, STING is subsequently phosphorylated by serine/threonine UNC-51-like kinase (ULK1/ATG1), and IRF3 function is suppressed. ULK1 activation occurred following disassociation from its repressor AMP activated protein kinase (AMPK) and was elicited by CDNs generated by the cGAMP synthase, cGAS. Thus, although CDNs may initially facilitate STING function, they subsequently trigger negative-feedback control of STING activity, thus preventing the persistent transcription of innate immune genes.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Taking the STING out of cytosolic DNA sensing.Trends Immunol. 2014 Jan;35(1):1-2. doi: 10.1016/j.it.2013.10.011. Epub 2013 Nov 11. Trends Immunol. 2014. PMID: 24231656
Similar articles
-
UV Light Potentiates STING (Stimulator of Interferon Genes)-dependent Innate Immune Signaling through Deregulation of ULK1 (Unc51-like Kinase 1).J Biol Chem. 2015 May 8;290(19):12184-94. doi: 10.1074/jbc.M115.649301. Epub 2015 Mar 19. J Biol Chem. 2015. PMID: 25792739 Free PMC article.
-
TBK1 and IKKε Act Redundantly to Mediate STING-Induced NF-κB Responses in Myeloid Cells.Cell Rep. 2020 Apr 7;31(1):107492. doi: 10.1016/j.celrep.2020.03.056. Cell Rep. 2020. PMID: 32268090
-
AMP-activated Kinase (AMPK) Promotes Innate Immunity and Antiviral Defense through Modulation of Stimulator of Interferon Genes (STING) Signaling.J Biol Chem. 2017 Jan 6;292(1):292-304. doi: 10.1074/jbc.M116.763268. Epub 2016 Nov 22. J Biol Chem. 2017. PMID: 27879319 Free PMC article.
-
The cGAS-STING signaling in cardiovascular and metabolic diseases: Future novel target option for pharmacotherapy.Acta Pharm Sin B. 2022 Jan;12(1):50-75. doi: 10.1016/j.apsb.2021.05.011. Epub 2021 May 20. Acta Pharm Sin B. 2022. PMID: 35127372 Free PMC article. Review.
-
Mechanisms and pathways of innate immune activation and regulation in health and cancer.Hum Vaccin Immunother. 2014;10(11):3270-85. doi: 10.4161/21645515.2014.979640. Hum Vaccin Immunother. 2014. PMID: 25625930 Free PMC article. Review.
Cited by
-
cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection.Proc Natl Acad Sci U S A. 2015 Apr 7;112(14):E1773-81. doi: 10.1073/pnas.1424637112. Epub 2015 Mar 23. Proc Natl Acad Sci U S A. 2015. PMID: 25831530 Free PMC article.
-
Innate immune pattern recognition: a cell biological perspective.Annu Rev Immunol. 2015;33:257-90. doi: 10.1146/annurev-immunol-032414-112240. Epub 2015 Jan 2. Annu Rev Immunol. 2015. PMID: 25581309 Free PMC article. Review.
-
New tricks for old dogs: countering antibiotic resistance in tuberculosis with host-directed therapeutics.Immunol Rev. 2015 Mar;264(1):344-62. doi: 10.1111/imr.12255. Immunol Rev. 2015. PMID: 25703571 Free PMC article. Review.
-
Interferon signaling in cancer. Non-canonical pathways and control of intracellular immune checkpoints.Semin Immunol. 2019 Jun;43:101299. doi: 10.1016/j.smim.2019.101299. Semin Immunol. 2019. PMID: 31771762 Free PMC article. Review.
-
Crosstalk Between Autophagy and the cGAS-STING Signaling Pathway in Type I Interferon Production.Front Cell Dev Biol. 2021 Nov 29;9:748485. doi: 10.3389/fcell.2021.748485. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34926445 Free PMC article. Review.
References
-
- Alexander A, Walker CL. The role of LKB1 and AMPK in cellular responses to stress and damage. FEBS Lett. 2011;585:952–957. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

