Beyond ubiquitination: the atypical functions of Fbxo7 and other F-box proteins
- PMID: 24107298
- PMCID: PMC3814724
- DOI: 10.1098/rsob.130131
Beyond ubiquitination: the atypical functions of Fbxo7 and other F-box proteins
Abstract
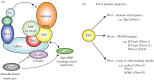
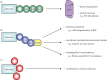
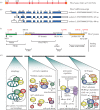
F-box proteins (FBPs) are substrate-recruiting subunits of Skp1-cullin1-FBP (SCF)-type E3 ubiquitin ligases. To date, 69 FBPs have been identified in humans, but ubiquitinated substrates have only been identified for a few, with the majority of FBPs remaining 'orphans'. In recent years, a growing body of work has identified non-canonical, SCF-independent roles for about 12% of the human FBPs. These atypical FBPs affect processes as diverse as transcription, cell cycle regulation, mitochondrial dynamics and intracellular trafficking. Here, we provide a general review of FBPs, with a particular emphasis on these expanded functions. We review Fbxo7 as an exemplar of this special group as it has well-defined roles in both SCF and non-SCF complexes. We review its function as a cell cycle regulator, via its ability to stabilize p27 protein and Cdk6 complexes, and as a proteasome regulator, owing to its high affinity binding to PI31. We also highlight recent advances in our understanding of Fbxo7 function in Parkinson's disease, where it functions in the regulation of mitophagy with PINK1 and Parkin. We postulate that a few extraordinary FBPs act as platforms that seamlessly segue their canonical and non-canonical functions to integrate different cellular pathways and link their regulation.
Keywords: E3 ligase; F-box protein; Fbxo7/PARK15; Parkinson's disease; mitophagy; ubiquitin.
Figures
Similar articles
-
Linking F-box protein 7 and parkin to neuronal degeneration in Parkinson's disease (PD).Mol Brain. 2016 Apr 18;9:41. doi: 10.1186/s13041-016-0218-2. Mol Brain. 2016. PMID: 27090516 Free PMC article. Review.
-
Structure and Function of Fbxo7/PARK15 in Parkinson's Disease.Curr Protein Pept Sci. 2017;18(7):715-724. doi: 10.2174/1389203717666160311121433. Curr Protein Pept Sci. 2017. PMID: 26965690 Review.
-
Gsk3β and Tomm20 are substrates of the SCFFbxo7/PARK15 ubiquitin ligase associated with Parkinson's disease.Biochem J. 2016 Oct 15;473(20):3563-3580. doi: 10.1042/BCJ20160387. Epub 2016 Aug 8. Biochem J. 2016. PMID: 27503909 Free PMC article.
-
A new mutation in the Parkinson's-related FBXO7 gene impairs mitochondrial and proteasomal function.FEBS J. 2024 Jun;291(12):2562-2564. doi: 10.1111/febs.17155. Epub 2024 May 6. FEBS J. 2024. PMID: 38708447
-
The E3 ubiquitin ligase SCF(Fbxo7) mediates proteasomal degradation of UXT isoform 2 (UXT-V2) to inhibit the NF-κB signaling pathway.Biochim Biophys Acta Gen Subj. 2021 Jan;1865(1):129754. doi: 10.1016/j.bbagen.2020.129754. Epub 2020 Sep 30. Biochim Biophys Acta Gen Subj. 2021. PMID: 33010352 Free PMC article.
Cited by
-
Mechanisms and roles of mitophagy in neurodegenerative diseases.CNS Neurosci Ther. 2019 Jul;25(7):859-875. doi: 10.1111/cns.13140. Epub 2019 May 2. CNS Neurosci Ther. 2019. PMID: 31050206 Free PMC article. Review.
-
Targeted inhibition of the COP9 signalosome for treatment of cancer.Nat Commun. 2016 Oct 24;7:13166. doi: 10.1038/ncomms13166. Nat Commun. 2016. PMID: 27774986 Free PMC article.
-
Functional characterization of Cullin-1-RING ubiquitin ligase (CRL1) complex in Leishmania infantum.PLoS Pathog. 2024 Jul 17;20(7):e1012336. doi: 10.1371/journal.ppat.1012336. eCollection 2024 Jul. PLoS Pathog. 2024. PMID: 39018347 Free PMC article.
-
Linking F-box protein 7 and parkin to neuronal degeneration in Parkinson's disease (PD).Mol Brain. 2016 Apr 18;9:41. doi: 10.1186/s13041-016-0218-2. Mol Brain. 2016. PMID: 27090516 Free PMC article. Review.
-
FBXO11 Mediates Ubiquitination of ZEB1 and Modulates Epithelial-to-Mesenchymal Transition in Lung Cancer Cells.Cancers (Basel). 2024 Sep 26;16(19):3269. doi: 10.3390/cancers16193269. Cancers (Basel). 2024. PMID: 39409891 Free PMC article.
References
-
- Reed SI. 2003. Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat. Rev. Mol. Cell Biol. 4, 855–864 (doi:10.1038/nrm1246) - DOI - PubMed
-
- Ang XL, Harper JW. 2004. Interwoven ubiquitination oscillators and control of cell cycle transitions. Sci. STKE 2004, pe31 (doi:10.1126/stke.2422004pe31) - DOI - PubMed
-
- Pickart CM. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 (doi:10.1146/annurev.biochem.70.1.503) - DOI - PubMed
-
- Pickart CM. 2004. Back to the future with ubiquitin. Cell 116, 181–190 (doi:10.1016/S0092-8674(03)01074-2) - DOI - PubMed
-
- Dye BT, Schulman BA. 2007. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu. Rev. Biophys. Biomol. Struct. 36, 131–150 (doi:10.1146/annurev.biophys.36.040306.132820) - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

