CAST-ChIP maps cell-type-specific chromatin states in the Drosophila central nervous system
- PMID: 24095734
- PMCID: PMC5877786
- DOI: 10.1016/j.celrep.2013.09.001
CAST-ChIP maps cell-type-specific chromatin states in the Drosophila central nervous system
Abstract
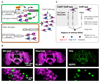
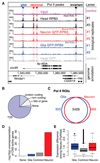
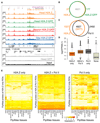
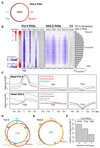
Chromatin organization and gene activity are responsive to developmental and environmental cues. Although many genes are transcribed throughout development and across cell types, much of gene regulation is highly cell-type specific. To readily track chromatin features at the resolution of cell types within complex tissues, we developed and validated chromatin affinity purification from specific cell types by chromatin immunoprecipitation (CAST-ChIP), a broadly applicable biochemical procedure. RNA polymerase II (Pol II) CAST-ChIP identifies ~1,500 neuronal and glia-specific genes in differentiated cells within the adult Drosophila brain. In contrast, the histone H2A.Z is distributed similarly across cell types and throughout development, marking cell-type-invariant Pol II-bound regions. Our study identifies H2A.Z as an active chromatin signature that is refractory to changes across cell fates. Thus, CAST-ChIP powerfully identifies cell-type-specific as well as cell-type-invariant chromatin states, enabling the systematic dissection of chromatin structure and gene regulation within complex tissues such as the brain.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Different chromatin interfaces of the Drosophila dosage compensation complex revealed by high-shear ChIP-seq.Genome Res. 2013 Mar;23(3):473-85. doi: 10.1101/gr.146407.112. Epub 2012 Dec 11. Genome Res. 2013. PMID: 23233545 Free PMC article.
-
Drosophila Paf1 modulates chromatin structure at actively transcribed genes.Mol Cell Biol. 2006 Jan;26(1):250-60. doi: 10.1128/MCB.26.1.250-260.2006. Mol Cell Biol. 2006. PMID: 16354696 Free PMC article.
-
Temporal ChIP-on-Chip of RNA-Polymerase-II to detect novel gene activation events during photoreceptor maturation.Mol Vis. 2010 Feb 17;16:252-71. Mol Vis. 2010. PMID: 20161818 Free PMC article.
-
Chromatin Immunoprecipitation for Analyzing Transcription Factor Binding and Histone Modifications in Drosophila.Methods Mol Biol. 2016;1478:263-277. doi: 10.1007/978-1-4939-6371-3_16. Methods Mol Biol. 2016. PMID: 27730588 Review.
-
Chromatin Architecture in the Fly: Living without CTCF/Cohesin Loop Extrusion?: Alternating Chromatin States Provide a Basis for Domain Architecture in Drosophila.Bioessays. 2019 Sep;41(9):e1900048. doi: 10.1002/bies.201900048. Epub 2019 Jul 1. Bioessays. 2019. PMID: 31264253 Review.
Cited by
-
Epigenetics and therapeutic targets mediating neuroprotection.Brain Res. 2015 Dec 2;1628(Pt B):265-272. doi: 10.1016/j.brainres.2015.07.034. Epub 2015 Jul 30. Brain Res. 2015. PMID: 26236020 Free PMC article. Review.
-
Studying alcohol use disorder using Drosophila melanogaster in the era of 'Big Data'.Behav Brain Funct. 2019 Apr 16;15(1):7. doi: 10.1186/s12993-019-0159-x. Behav Brain Funct. 2019. PMID: 30992041 Free PMC article. Review.
-
DamID profiling of dynamic Polycomb-binding sites in Drosophila imaginal disc development and tumorigenesis.Epigenetics Chromatin. 2018 Jun 5;11(1):27. doi: 10.1186/s13072-018-0196-y. Epigenetics Chromatin. 2018. PMID: 29871666 Free PMC article.
-
High-resolution digital profiling of the epigenome.Nat Rev Genet. 2014 Dec;15(12):814-27. doi: 10.1038/nrg3798. Epub 2014 Oct 9. Nat Rev Genet. 2014. PMID: 25297728 Review.
-
Histone H2A.Z subunit exchange controls consolidation of recent and remote memory.Nature. 2014 Nov 27;515(7528):582-6. doi: 10.1038/nature13707. Epub 2014 Sep 14. Nature. 2014. PMID: 25219850 Free PMC article.
References
-
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczyński B, Riddell A, Furlong EEM. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–156. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

