Archaeal signal transduction: impact of protein phosphatase deletions on cell size, motility, and energy metabolism in Sulfolobus acidocaldarius
- PMID: 24078887
- PMCID: PMC3861733
- DOI: 10.1074/mcp.M113.027375
Archaeal signal transduction: impact of protein phosphatase deletions on cell size, motility, and energy metabolism in Sulfolobus acidocaldarius
Abstract
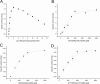
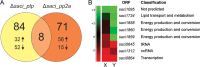
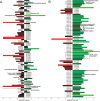
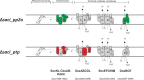
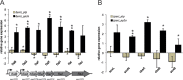
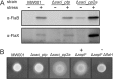
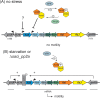
In this study, the in vitro and in vivo functions of the only two identified protein phosphatases, Saci-PTP and Saci-PP2A, in the crenarchaeal model organism Sulfolobus acidocaldarius were investigated. Biochemical characterization revealed that Saci-PTP is a dual-specific phosphatase (against pSer/pThr and pTyr), whereas Saci-PP2A exhibited specific pSer/pThr activity and inhibition by okadaic acid. Deletion of saci_pp2a resulted in pronounced alterations in growth, cell shape and cell size, which could be partially complemented. Transcriptome analysis of the three strains (Δsaci_ptp, Δsaci_pp2a and the MW001 parental strain) revealed 155 genes that were differentially expressed in the deletion mutants, and showed significant changes in expression of genes encoding the archaella (archaeal motility structure), components of the respiratory chain and transcriptional regulators. Phosphoproteome studies revealed 801 unique phosphoproteins in total, with an increase in identified phosphopeptides in the deletion mutants. Proteins from most functional categories were affected by phosphorylation, including components of the motility system, the respiratory chain, and regulatory proteins. In the saci_pp2a deletion mutant the up-regulation at the transcript level, as well as the observed phosphorylation pattern, resembled starvation stress responses. Hypermotility was also observed in the saci_pp2a deletion mutant. The results highlight the importance of protein phosphorylation in regulating essential cellular processes in the crenarchaeon S. acidocaldarius.
Figures

Similar articles
-
Expanding the archaellum regulatory network - the eukaryotic protein kinases ArnC and ArnD influence motility of Sulfolobus acidocaldarius.Microbiologyopen. 2017 Feb;6(1):e00414. doi: 10.1002/mbo3.414. Epub 2016 Oct 22. Microbiologyopen. 2017. PMID: 27771939 Free PMC article.
-
Sulfolobus acidocaldarius Transports Pentoses via a Carbohydrate Uptake Transporter 2 (CUT2)-Type ABC Transporter and Metabolizes Them through the Aldolase-Independent Weimberg Pathway.Appl Environ Microbiol. 2018 Jan 17;84(3):e01273-17. doi: 10.1128/AEM.01273-17. Print 2018 Feb 1. Appl Environ Microbiol. 2018. PMID: 29150511 Free PMC article.
-
Alterations of the transcriptome of Sulfolobus acidocaldarius by exoribonuclease aCPSF2.PLoS One. 2013 Oct 7;8(10):e76569. doi: 10.1371/journal.pone.0076569. eCollection 2013. PLoS One. 2013. PMID: 24116119 Free PMC article.
-
Evolutionary crossroads of cell signaling: PP1 and PP2A substrate sites in intrinsically disordered regions.Biochem Soc Trans. 2021 Jun 30;49(3):1065-1074. doi: 10.1042/BST20200175. Biochem Soc Trans. 2021. PMID: 34100859 Free PMC article. Review.
-
Homologous recombination in Sulfolobus acidocaldarius: genetic assays and functional properties.Biochem Soc Trans. 2009 Feb;37(Pt 1):88-91. doi: 10.1042/BST0370088. Biochem Soc Trans. 2009. PMID: 19143608 Review.
Cited by
-
The Phosphatase PP2A Interacts With ArnA and ArnB to Regulate the Oligomeric State and the Stability of the ArnA/B Complex.Front Microbiol. 2020 Aug 21;11:1849. doi: 10.3389/fmicb.2020.01849. eCollection 2020. Front Microbiol. 2020. PMID: 32973695 Free PMC article.
-
Structure and interactions of the archaeal motility repression module ArnA-ArnB that modulates archaellum gene expression in Sulfolobus acidocaldarius.J Biol Chem. 2019 May 3;294(18):7460-7471. doi: 10.1074/jbc.RA119.007709. Epub 2019 Mar 22. J Biol Chem. 2019. PMID: 30902813 Free PMC article.
-
Carbohydrate metabolism in Archaea: current insights into unusual enzymes and pathways and their regulation.Microbiol Mol Biol Rev. 2014 Mar;78(1):89-175. doi: 10.1128/MMBR.00041-13. Microbiol Mol Biol Rev. 2014. PMID: 24600042 Free PMC article. Review.
-
The interplay between nucleoid organization and transcription in archaeal genomes.Nat Rev Microbiol. 2015 Jun;13(6):333-41. doi: 10.1038/nrmicro3467. Epub 2015 May 6. Nat Rev Microbiol. 2015. PMID: 25944489 Review.
-
The Impact of Pyroglutamate: Sulfolobus acidocaldarius Has a Growth Advantage over Saccharolobus solfataricus in Glutamate-Containing Media.Archaea. 2019 Apr 24;2019:3208051. doi: 10.1155/2019/3208051. eCollection 2019. Archaea. 2019. PMID: 31178666 Free PMC article.
References
-
- Macek B., Mijakovic I., Olsen J. V., Gnad F., Kumar C., Jensen P. R., Mann M. (2007) The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6, 697–707 - PubMed
-
- Pawson T., Scott J. D. (2005) Protein phosphorylation in signaling - 50 years and counting. Trends Biochem. Sci. 30, 286–290 - PubMed
-
- Johnson L. N., Lewis R. J. (2001) Structural basis for control by phosphorylation. Chem. Rev. 101, 2209–2242 - PubMed
-
- Westheimer F. H. (1987) Why nature chose phosphates. Science 235, 1173–1178 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

