Alternative spliceosome assembly pathways revealed by single-molecule fluorescence microscopy
- PMID: 24075986
- PMCID: PMC3927372
- DOI: 10.1016/j.celrep.2013.08.026
Alternative spliceosome assembly pathways revealed by single-molecule fluorescence microscopy
Abstract
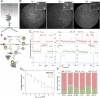
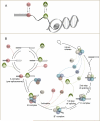
Removal of introns from nascent transcripts (pre-mRNAs) by the spliceosome is an essential step in eukaryotic gene expression. Previous studies have suggested that the earliest steps in spliceosome assembly in yeast are highly ordered and the stable recruitment of U1 small nuclear ribonucleoprotein particle (snRNP) to the 5' splice site necessarily precedes recruitment of U2 snRNP to the branch site to form the "prespliceosome." Here, using colocalization single-molecule spectroscopy to follow initial spliceosome assembly on eight different S. cerevisiae pre-mRNAs, we demonstrate that active yeast spliceosomes can form by both U1-first and U2-first pathways. Both assembly pathways yield prespliceosomes functionally equivalent for subsequent U5·U4/U6 tri-snRNP recruitment and for intron excision. Although fractional flux through the two pathways varies on different introns, both are operational on all introns studied. Thus, multiple pathways exist for assembling functional spliceosomes. These observations provide insight into the mechanisms of cross-intron coordination of initial spliceosome assembly.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Single molecule analysis reveals reversible and irreversible steps during spliceosome activation.Elife. 2016 May 31;5:e14166. doi: 10.7554/eLife.14166. Elife. 2016. PMID: 27244240 Free PMC article.
-
Prespliceosome structure provides insights into spliceosome assembly and regulation.Nature. 2018 Jul;559(7714):419-422. doi: 10.1038/s41586-018-0323-8. Epub 2018 Jul 11. Nature. 2018. PMID: 29995849 Free PMC article.
-
Structure of a pre-catalytic spliceosome.Nature. 2017 Jun 29;546(7660):617-621. doi: 10.1038/nature22799. Epub 2017 May 22. Nature. 2017. PMID: 28530653 Free PMC article.
-
RNA Splicing by the Spliceosome.Annu Rev Biochem. 2020 Jun 20;89:359-388. doi: 10.1146/annurev-biochem-091719-064225. Epub 2019 Dec 3. Annu Rev Biochem. 2020. PMID: 31794245 Review.
-
How Is Precursor Messenger RNA Spliced by the Spliceosome?Annu Rev Biochem. 2020 Jun 20;89:333-358. doi: 10.1146/annurev-biochem-013118-111024. Epub 2019 Dec 9. Annu Rev Biochem. 2020. PMID: 31815536 Review.
Cited by
-
Single molecule analysis reveals reversible and irreversible steps during spliceosome activation.Elife. 2016 May 31;5:e14166. doi: 10.7554/eLife.14166. Elife. 2016. PMID: 27244240 Free PMC article.
-
Transcription Increases the Cooperativity of Ribonucleoprotein Assembly.Cell. 2019 Nov 27;179(6):1370-1381.e12. doi: 10.1016/j.cell.2019.11.007. Epub 2019 Nov 21. Cell. 2019. PMID: 31761536 Free PMC article.
-
Splicing heterogeneity: separating signal from noise.Genome Biol. 2018 Jul 9;19(1):86. doi: 10.1186/s13059-018-1467-4. Genome Biol. 2018. PMID: 29986741 Free PMC article.
-
Dynamics and consequences of spliceosome E complex formation.Elife. 2017 Aug 22;6:e27592. doi: 10.7554/eLife.27592. Elife. 2017. PMID: 28829039 Free PMC article.
-
What have single-molecule studies taught us about gene expression?Genes Dev. 2016 Aug 15;30(16):1796-810. doi: 10.1101/gad.281725.116. Genes Dev. 2016. PMID: 27601529 Free PMC article. Review.
References
-
- Crispino JD, Blencowe BJ, Sharp PA. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science. 1994;265:1866–1869. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

