Structural basis for the divergent evolution of influenza B virus hemagglutinin
- PMID: 24074573
- PMCID: PMC3902124
- DOI: 10.1016/j.virol.2013.07.035
Structural basis for the divergent evolution of influenza B virus hemagglutinin
Abstract
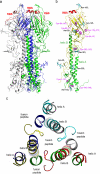
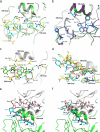
Influenza A and B viruses are responsible for the severe morbidity and mortality worldwide in annual influenza epidemics. Currently circulating influenza B virus belongs to the B/Victoria or B/Yamagata lineage that was diverged from each other about 30-40 years ago. However, a mechanistic understanding of their divergent evolution is still lacking. Here we report the crystal structures of influenza B/Yamanashi/166/1998 hemagglutinin (HA) belonging to B/Yamagata lineage and its complex with the avian-like receptor analogue. Comparison of these structures with those of undiverged and diverged influenza B virus HAs, in conjunction with sequence analysis, reveals the molecular basis for the divergent evolution of influenza B virus HAs. Furthermore, HAs of diverged influenza B virus strains display much stronger molecular interactions with terminal sialic acid of bound receptors, which may allow for a different tissue tropism for current influenza B viruses, for which further investigation is required.
Keywords: Divergent evolution; Hemagglutinin; Influenza B virus; Positive selective pressure; Receptor binding; Sialic acid receptors.
© 2013 Elsevier Inc. All rights reserved.
Figures
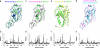

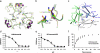
Similar articles
-
Influenza B Virus Receptor Specificity: Closing the Gap between Binding and Tropism.Viruses. 2024 Aug 24;16(9):1356. doi: 10.3390/v16091356. Viruses. 2024. PMID: 39339833 Free PMC article. Review.
-
Structure and receptor binding of the hemagglutinin from a human H6N1 influenza virus.Cell Host Microbe. 2015 Mar 11;17(3):369-376. doi: 10.1016/j.chom.2015.02.005. Cell Host Microbe. 2015. PMID: 25766295 Free PMC article.
-
Structure and receptor binding specificity of hemagglutinin H13 from avian influenza A virus H13N6.J Virol. 2013 Aug;87(16):9077-85. doi: 10.1128/JVI.00235-13. Epub 2013 Jun 12. J Virol. 2013. PMID: 23760233 Free PMC article.
-
Molecular characterization of the receptor binding structure-activity relationships of influenza B virus hemagglutinin.Acta Virol. 2013;57(3):313-32. Acta Virol. 2013. PMID: 24020757
-
[The Victoria and Yamagata Lineages of Influenza B Viruses, unknown and undervalued].Rev Esp Quimioter. 2022 Jun;35(3):231-235. doi: 10.37201/req/159.2021. Epub 2022 Feb 18. Rev Esp Quimioter. 2022. PMID: 35180825 Free PMC article. Review. Spanish.
Cited by
-
Influenza B Virus Receptor Specificity: Closing the Gap between Binding and Tropism.Viruses. 2024 Aug 24;16(9):1356. doi: 10.3390/v16091356. Viruses. 2024. PMID: 39339833 Free PMC article. Review.
-
Correlation of Influenza B Haemagglutination Inhibiton, Single-Radial Haemolysis and Pseudotype-Based Microneutralisation Assays for Immunogenicity Testing of Seasonal Vaccines.Vaccines (Basel). 2021 Jan 28;9(2):100. doi: 10.3390/vaccines9020100. Vaccines (Basel). 2021. PMID: 33525543 Free PMC article.
-
Epidemiological, Clinical and Virological Characteristics of Influenza B Virus from Patients at the Hospital Tertiary Care Units in Bangkok during 2011-2014.PLoS One. 2016 Jul 7;11(7):e0158244. doi: 10.1371/journal.pone.0158244. eCollection 2016. PLoS One. 2016. PMID: 27387488 Free PMC article.
-
Hemagglutinin Cleavability, Acid Stability, and Temperature Dependence Optimize Influenza B Virus for Replication in Human Airways.J Virol. 2019 Dec 12;94(1):e01430-19. doi: 10.1128/JVI.01430-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597759 Free PMC article.
-
Molecular evolution of influenza B virus during 2011-2017 in Chaoyang, Beijing, suggesting the free influenza vaccine policy.Sci Rep. 2019 Feb 21;9(1):2432. doi: 10.1038/s41598-018-38105-1. Sci Rep. 2019. PMID: 30792414 Free PMC article.
References
-
- Abed Y, Coulthart MB, Li Y, Boivin G. Evolution of surface and nonstructural-1 genes of influenza B viruses isolated in the Province of Quebec, Canada, during the 1998–2001 period. Virus Genes. 2003;27(2):125–135. - PubMed
-
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66(2):213–221. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

