Architecture of an RNA polymerase II transcription pre-initiation complex
- PMID: 24072820
- PMCID: PMC4039082
- DOI: 10.1126/science.1238724
Architecture of an RNA polymerase II transcription pre-initiation complex
Abstract
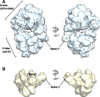
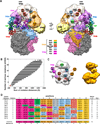
The protein density and arrangement of subunits of a complete, 32-protein, RNA polymerase II (pol II) transcription pre-initiation complex (PIC) were determined by means of cryogenic electron microscopy and a combination of chemical cross-linking and mass spectrometry. The PIC showed a marked division in two parts, one containing all the general transcription factors (GTFs) and the other pol II. Promoter DNA was associated only with the GTFs, suspended above the pol II cleft and not in contact with pol II. This structural principle of the PIC underlies its conversion to a transcriptionally active state; the PIC is poised for the formation of a transcription bubble and descent of the DNA into the pol II cleft.
Figures



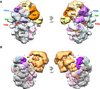
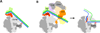
Similar articles
-
Promoter Distortion and Opening in the RNA Polymerase II Cleft.Mol Cell. 2019 Jan 3;73(1):97-106.e4. doi: 10.1016/j.molcel.2018.10.014. Epub 2018 Nov 21. Mol Cell. 2019. PMID: 30472190
-
Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.Mol Cell. 2022 Feb 3;82(3):660-676.e9. doi: 10.1016/j.molcel.2021.12.020. Epub 2022 Jan 19. Mol Cell. 2022. PMID: 35051353 Free PMC article.
-
Structural basis of RNA polymerase III transcription initiation.Nature. 2018 Jan 17;553(7688):301-306. doi: 10.1038/nature25441. Nature. 2018. PMID: 29345637
-
More pieces to the puzzle: recent structural insights into class II transcription initiation.Curr Opin Struct Biol. 2014 Feb;24:91-7. doi: 10.1016/j.sbi.2013.12.005. Epub 2014 Jan 16. Curr Opin Struct Biol. 2014. PMID: 24440461 Review.
-
Structural basis of transcription initiation by RNA polymerase II.Nat Rev Mol Cell Biol. 2015 Mar;16(3):129-43. doi: 10.1038/nrm3952. Epub 2015 Feb 18. Nat Rev Mol Cell Biol. 2015. PMID: 25693126 Review.
Cited by
-
A Conserved Nuclear Cyclophilin Is Required for Both RNA Polymerase II Elongation and Co-transcriptional Splicing in Caenorhabditis elegans.PLoS Genet. 2016 Aug 19;12(8):e1006227. doi: 10.1371/journal.pgen.1006227. eCollection 2016 Aug. PLoS Genet. 2016. PMID: 27541139 Free PMC article.
-
Epigenetic changes in the developing brain: Effects on behavior.Proc Natl Acad Sci U S A. 2015 Jun 2;112(22):6789-95. doi: 10.1073/pnas.1501482112. Proc Natl Acad Sci U S A. 2015. PMID: 26034282 Free PMC article. No abstract available.
-
Gdown1 Associates Efficiently with RNA Polymerase II after Promoter Clearance and Displaces TFIIF during Transcript Elongation.PLoS One. 2016 Oct 7;11(10):e0163649. doi: 10.1371/journal.pone.0163649. eCollection 2016. PLoS One. 2016. PMID: 27716820 Free PMC article.
-
RNAs nonspecifically inhibit RNA polymerase II by preventing binding to the DNA template.RNA. 2014 May;20(5):644-55. doi: 10.1261/rna.040444.113. Epub 2014 Mar 10. RNA. 2014. PMID: 24614752 Free PMC article.
-
In TFIIH, XPD helicase is exclusively devoted to DNA repair.PLoS Biol. 2014 Sep 30;12(9):e1001954. doi: 10.1371/journal.pbio.1001954. eCollection 2014 Sep. PLoS Biol. 2014. PMID: 25268380 Free PMC article.
References
-
- Conaway RC, Conaway JW. General initiation factors for RNA polymerase II. Annu Rev Biochem. 1993;62:161. - PubMed
-
- Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005 May;30:235. - PubMed
-
- Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769. - PubMed
-
- Buratowski S, Hahn S, Guarente L, Sharp PA. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989 Feb 24;56:549. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

