N-alpha-terminal acetylation of histone H4 regulates arginine methylation and ribosomal DNA silencing
- PMID: 24068969
- PMCID: PMC3778019
- DOI: 10.1371/journal.pgen.1003805
N-alpha-terminal acetylation of histone H4 regulates arginine methylation and ribosomal DNA silencing
Abstract
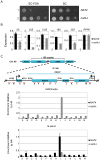
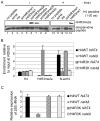
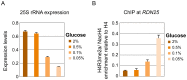
Post-translational modifications of histones play a key role in DNA-based processes, like transcription, by modulating chromatin structure. N-terminal acetylation is unique among the numerous histone modifications because it is deposited on the N-alpha amino group of the first residue instead of the side-chain of amino acids. The function of this modification and its interplay with other internal histone marks has not been previously addressed. Here, we identified N-terminal acetylation of H4 (N-acH4) as a novel regulator of arginine methylation and chromatin silencing in Saccharomyces cerevisiae. Lack of the H4 N-alpha acetyltransferase (Nat4) activity results specifically in increased deposition of asymmetric dimethylation of histone H4 arginine 3 (H4R3me2a) and in enhanced ribosomal-DNA silencing. Consistent with this, H4 N-terminal acetylation impairs the activity of the Hmt1 methyltransferase towards H4R3 in vitro. Furthermore, combinatorial loss of N-acH4 with internal histone acetylation at lysines 5, 8 and 12 has a synergistic induction of H4R3me2a deposition and rDNA silencing that leads to a severe growth defect. This defect is completely rescued by mutating arginine 3 to lysine (H4R3K), suggesting that abnormal deposition of a single histone modification, H4R3me2a, can impact on cell growth. Notably, the cross-talk between N-acH4 and H4R3me2a, which regulates rDNA silencing, is induced under calorie restriction conditions. Collectively, these findings unveil a molecular and biological function for H4 N-terminal acetylation, identify its interplay with internal histone modifications, and provide general mechanistic implications for N-alpha-terminal acetylation, one of the most common protein modifications in eukaryotes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Loss of Nat4 and its associated histone H4 N-terminal acetylation mediates calorie restriction-induced longevity.EMBO Rep. 2016 Dec;17(12):1829-1843. doi: 10.15252/embr.201642540. Epub 2016 Oct 31. EMBO Rep. 2016. PMID: 27799288 Free PMC article.
-
The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae.Genes Dev. 2001 Dec 1;15(23):3169-82. doi: 10.1101/gad.929001. Genes Dev. 2001. PMID: 11731480 Free PMC article.
-
Histone H4 lysine 16 acetylation regulates cellular lifespan.Nature. 2009 Jun 11;459(7248):802-7. doi: 10.1038/nature08085. Nature. 2009. PMID: 19516333 Free PMC article.
-
Histone post-translational modifications regulate transcription and silent chromatin in Saccharomyces cerevisiae.Ernst Schering Res Found Workshop. 2006;(57):127-53. doi: 10.1007/3-540-37633-x_8. Ernst Schering Res Found Workshop. 2006. PMID: 16568953 Review.
-
Chatting histone modifications in mammals.Brief Funct Genomics. 2010 Dec;9(5-6):429-43. doi: 10.1093/bfgp/elq024. Brief Funct Genomics. 2010. PMID: 21266346 Free PMC article. Review.
Cited by
-
Probing the interaction between NatA and the ribosome for co-translational protein acetylation.PLoS One. 2017 Oct 10;12(10):e0186278. doi: 10.1371/journal.pone.0186278. eCollection 2017. PLoS One. 2017. PMID: 29016658 Free PMC article.
-
Nucleolar and Ribosomal DNA Structure under Stress: Yeast Lessons for Aging and Cancer.Cells. 2019 Jul 26;8(8):779. doi: 10.3390/cells8080779. Cells. 2019. PMID: 31357498 Free PMC article. Review.
-
Aging and Caloric Restriction Modulate the DNA Methylation Profile of the Ribosomal RNA Locus in Human and Rat Liver.Nutrients. 2020 Jan 21;12(2):277. doi: 10.3390/nu12020277. Nutrients. 2020. PMID: 31973116 Free PMC article.
-
From Nucleus to Membrane: A Subcellular Map of the N-Acetylation Machinery in Plants.Int J Mol Sci. 2022 Nov 21;23(22):14492. doi: 10.3390/ijms232214492. Int J Mol Sci. 2022. PMID: 36430970 Free PMC article. Review.
-
N-terminal acetylation of Set1-COMPASS fine-tunes H3K4 methylation patterns.Sci Adv. 2024 Jul 12;10(28):eadl6280. doi: 10.1126/sciadv.adl6280. Epub 2024 Jul 12. Sci Adv. 2024. PMID: 38996018 Free PMC article.
References
-
- Brown JL, Roberts WK (1976) Evidence that approximately eighty per cent of the soluble proteins from Ehrlich ascites cells are Nalpha-acetylated. J Biol Chem 251: 1009–1014. - PubMed
-
- Starheim KK, Gevaert K, Arnesen T (2012) Protein N-terminal acetyltransferases: when the start matters. Trends Biochem Sci 37: 152–161. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

