Evolution of BRET Biosensors from Live Cell to Tissue-Scale In vivo Imaging
- PMID: 24065957
- PMCID: PMC3779814
- DOI: 10.3389/fendo.2013.00131
Evolution of BRET Biosensors from Live Cell to Tissue-Scale In vivo Imaging
Abstract
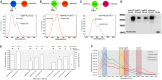
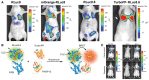
Development of bioluminescence resonance energy transfer (BRET) based genetic sensors for sensing biological functions such as protein-protein interactions (PPIs) in vivo has a special value in measuring such dynamic events at their native environment. Since its inception in the late nineties, BRET related research has gained significant momentum in terms of adding versatility to the assay format and wider applicability where it has been suitably used. Beyond the scope of quantitative measurement of PPIs and protein dimerization, molecular imaging applications based on BRET assays have broadened its scope for screening pharmacologically important compounds by in vivo imaging as well. In this mini-review we focus on an in-depth analysis of engineered BRET systems developed and their successful application to cell-based assays as well as in vivo non-invasive imaging in live subjects.
Keywords: bioluminescence resonance energy transfer; cell-based assay; fluorescent proteins; luciferase; optical imaging; protein–protein interactions.
Figures
Similar articles
-
Use of BRET to Study Protein-Protein Interactions In Vitro and In Vivo.Methods Mol Biol. 2016;1443:57-78. doi: 10.1007/978-1-4939-3724-0_5. Methods Mol Biol. 2016. PMID: 27246334
-
Reporter-Based BRET Sensors for Measuring Biological Functions In Vivo.Methods Mol Biol. 2018;1790:51-74. doi: 10.1007/978-1-4939-7860-1_5. Methods Mol Biol. 2018. PMID: 29858783
-
The new era of bioluminescence resonance energy transfer technology.Curr Pharm Biotechnol. 2011 Apr;12(4):558-68. doi: 10.2174/138920111795163922. Curr Pharm Biotechnol. 2011. PMID: 21342101 Review.
-
In Vivo Assessment of Protein-Protein Interactions Using BRET Assay.Methods Mol Biol. 2022;2525:239-257. doi: 10.1007/978-1-0716-2473-9_18. Methods Mol Biol. 2022. PMID: 35836073
-
NanoBRET: The Bright Future of Proximity-Based Assays.Front Bioeng Biotechnol. 2019 Mar 26;7:56. doi: 10.3389/fbioe.2019.00056. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 30972335 Free PMC article. Review.
Cited by
-
Comment on "The Use of BRET to Study Receptor-Protein Interactions".Front Endocrinol (Lausanne). 2014 Jan 22;5:3. doi: 10.3389/fendo.2014.00003. eCollection 2014. Front Endocrinol (Lausanne). 2014. PMID: 24478757 Free PMC article. No abstract available.
-
Bioluminescence imaging in live cells and animals.Neurophotonics. 2016 Apr;3(2):025001. doi: 10.1117/1.NPh.3.2.025001. Epub 2016 Apr 5. Neurophotonics. 2016. PMID: 27226972 Free PMC article.
-
Novel BRET combination for detection of rapamycin-induced protein dimerization using luciferase from fungus Neonothopanusnambi.Heliyon. 2024 Feb 8;10(4):e25553. doi: 10.1016/j.heliyon.2024.e25553. eCollection 2024 Feb 29. Heliyon. 2024. PMID: 38384550 Free PMC article.
-
In vivo opioid receptor heteromerization: where do we stand?Br J Pharmacol. 2015 Jan;172(2):420-34. doi: 10.1111/bph.12702. Epub 2014 Jul 1. Br J Pharmacol. 2015. PMID: 24666391 Free PMC article. Review.
-
In Vivo Analysis of Protein-Protein Interactions with Bioluminescence Resonance Energy Transfer (BRET): Progress and Prospects.Int J Mol Sci. 2016 Oct 11;17(10):1704. doi: 10.3390/ijms17101704. Int J Mol Sci. 2016. PMID: 27727181 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

