Characterization of the Stoichiometry of HMGA1/DNA Complexes
- PMID: 24062859
- PMCID: PMC3778555
- DOI: 10.2174/1874091X01307010073
Characterization of the Stoichiometry of HMGA1/DNA Complexes
Abstract
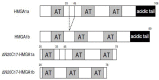
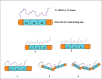
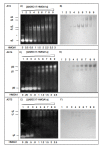
High-mobility group A1 (HMGA1) non-histone chromatin architectural transcription factors regulate gene expression, embryogenesis, cell differentiation, and adaptive immune responses by binding DNA and other transcription factors. HMGA1 has also been shown to be highly over-expressed in many human cancers and is considered to be a valuable cancer biomarker. Elevated HMGA1 expression levels also make cancer cells resistant to chemotherapy. Here, HMGA1/DNA complex formation was investigated using electrophoretic mobility shift assays (EMSA). Collectively, the EMSA results indicated that full length HMGA1 mixed with DNA containing three AT-hook binding sites formed four distinct HMGA1/DNA complexes ranging in stoichiometry from 1:2 to 3:1 in HMGA1:DNA ratio. The data indicated that the distribution of complexes with different HMGA1 to DNA stoichiometries depended on the molar ratio of HMGA1 to DNA in solution, which could have significant biological implications given that HMGA1 is highly over-expressed in human cancer cells. The two naturally occurring isoforms of HMGA1, HMGA1a and HMGA1b, the latter containing an 11 amino acid deletion between the first and second AT-hooks, were observed to have slightly different DNA binding profiles. Finally, HMGA1 binding affinity to DNA was found to be influenced by the DNA A:T segment sequence context, with higher specificity be observed in HMGA1 binding to TnAn segments, which have two local minor groove minima on either side of the TpA step, compared to An:Tn segments, which have a single minor groove minimum at the 3' end of the An run, implying AT-hook binding favors narrow minor groove structure.
Keywords: AT-hook; DNA.; EMSA; HMGA1; architectural transcription factor; enhanceosome.
Figures

Similar articles
-
A mass spectrometric study on the in vitro methylation of HMGA1a and HMGA1b proteins by PRMTs: methylation specificity, the effect of binding to AT-rich duplex DNA, and the effect of C-terminal phosphorylation.Biochemistry. 2007 Jul 3;46(26):7896-906. doi: 10.1021/bi6024897. Epub 2007 Jun 6. Biochemistry. 2007. PMID: 17550233
-
High mobility group A1 and cancer: potential biomarker and therapeutic target.Histol Histopathol. 2012 May;27(5):567-79. doi: 10.14670/HH-27.567. Histol Histopathol. 2012. PMID: 22419021 Review.
-
HMGA1 down-regulation is crucial for chromatin composition and a gene expression profile permitting myogenic differentiation.BMC Cell Biol. 2010 Aug 11;11:64. doi: 10.1186/1471-2121-11-64. BMC Cell Biol. 2010. PMID: 20701767 Free PMC article.
-
Netropsin improves survival from endotoxaemia by disrupting HMGA1 binding to the NOS2 promoter.Biochem J. 2009 Feb 15;418(1):103-12. doi: 10.1042/BJ20081427. Biochem J. 2009. PMID: 18937643 Free PMC article.
-
The High Mobility Group A1 (HMGA1) Transcriptome in Cancer and Development.Curr Mol Med. 2016;16(4):353-93. doi: 10.2174/1566524016666160316152147. Curr Mol Med. 2016. PMID: 26980699 Free PMC article. Review.
Cited by
-
Binding of high mobility group A proteins to the mammalian genome occurs as a function of AT-content.PLoS Genet. 2017 Dec 21;13(12):e1007102. doi: 10.1371/journal.pgen.1007102. eCollection 2017 Dec. PLoS Genet. 2017. PMID: 29267285 Free PMC article.
-
High Mobility Group A1 (HMGA1): Structure, Biological Function, and Therapeutic Potential.Int J Biol Sci. 2022 Jul 4;18(11):4414-4431. doi: 10.7150/ijbs.72952. eCollection 2022. Int J Biol Sci. 2022. PMID: 35864955 Free PMC article. Review.
-
Adenovirus-Mediated Delivery of Decoy Hyper Binding Sites Targeting Oncogenic HMGA1 Reduces Pancreatic and Liver Cancer Cell Viability.Mol Ther Oncolytics. 2018 Jan 31;8:52-61. doi: 10.1016/j.omto.2018.01.002. eCollection 2018 Mar 30. Mol Ther Oncolytics. 2018. PMID: 29511732 Free PMC article.
-
A mouse model study of toxicity and biodistribution of a replication defective adenovirus serotype 5 virus with its genome engineered to contain a decoy hyper binding site to sequester and suppress oncogenic HMGA1 as a new cancer treatment therapy.PLoS One. 2018 Feb 20;13(2):e0192882. doi: 10.1371/journal.pone.0192882. eCollection 2018. PLoS One. 2018. PMID: 29462157 Free PMC article.
-
HMGA1 regulates trabectedin sensitivity in advanced soft-tissue sarcoma (STS): A Spanish Group for Research on Sarcomas (GEIS) study.Cell Mol Life Sci. 2024 May 17;81(1):219. doi: 10.1007/s00018-024-05250-y. Cell Mol Life Sci. 2024. PMID: 38758230 Free PMC article.
References
-
- Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. - PubMed
-
- Murua Escobar H, Soller J T, Richter A, Meyer B, Winkler S, Bullerdiek J, Nolte I. "Best friends" sharing the HMGA1 gene: comparison of the human and canine HMGA1 to orthologous other species. J. Hered. 2005;96:777–781. - PubMed
-
- Bustin M. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci. 2001;26:152–153. - PubMed
-
- Chiappetta G, Avantaggiato V, Visconti R, Fedele M, Battista S, Trapasso F, Merciai B M, Fidanza V, Giancotti V, Santoro M, Simeone A, Fusco A. High level expression of the HMGI (Y) gene during embryonic development. Oncogene. 1996;13:2439–2446. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
