Etiology of diarrhea in Bangladeshi infants in the first year of life analyzed using molecular methods
- PMID: 24041797
- PMCID: PMC3814844
- DOI: 10.1093/infdis/jit507
Etiology of diarrhea in Bangladeshi infants in the first year of life analyzed using molecular methods
Abstract
Background: Diarrhea causes enormous morbidity and mortality in developing countries, yet the relative importance of multiple potential enteropathogens has been difficult to ascertain.
Methods: We performed a longitudinal cohort study from birth to 1 year of age in 147 infants in Dhaka, Bangladesh. Using multiplex polymerase chain reaction, we analyzed 420 episodes of diarrhea and 1385 monthly surveillance stool specimens for 32 enteropathogen gene targets. For each infant we examined enteropathogen quantities over time to ascribe each positive target as a probable or less-likely contributor to diarrhea.
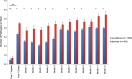
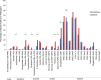
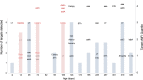
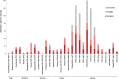
Results: Multiple enteropathogens were detected by the first month of life. Diarrhea was associated with a state of overall pathogen excess (mean number of enteropathogen gene targets (± SE), 5.6 ± 0.1 vs 4.3 ± 0.1 in surveillance stool specimens; P < .05). After a longitudinal, quantitative approach was applied to filter out less-likely contributors, each diarrheal episode still had an average of 3.3 probable or dominant targets. Enteroaggregative Escherichia coli, Campylobacter, enteropathogenic E. coli, rotavirus, and Entamoeba histolytica were the most frequent probable contributors to diarrhea. Rotavirus was enriched in moderate to severe diarrheal episodes.
Conclusions: In this community-based study diarrhea seemed to be a multipathogen event and a state of enteropathogen excess above a high carriage baseline.
Keywords: Campylobacter; Diarrhea; PCR; enteroaggregative e.coli; rotavirus.
Figures

Comment in
-
The intestinal pathobiome: its reality and consequences among infants and young children in resource-limited settings.J Infect Dis. 2013 Dec 1;208(11):1732-3. doi: 10.1093/infdis/jit509. Epub 2013 Sep 16. J Infect Dis. 2013. PMID: 24041794 No abstract available.
Similar articles
-
Etiology of childhood diarrhea after rotavirus vaccine introduction: a prospective, population-based study in Nicaragua.Pediatr Infect Dis J. 2014 Nov;33(11):1156-63. doi: 10.1097/INF.0000000000000427. Pediatr Infect Dis J. 2014. PMID: 24879131 Free PMC article.
-
Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago, Chile.Am J Epidemiol. 1993 Nov 15;138(10):849-69. doi: 10.1093/oxfordjournals.aje.a116788. Am J Epidemiol. 1993. PMID: 8237973
-
The intestinal pathobiome: its reality and consequences among infants and young children in resource-limited settings.J Infect Dis. 2013 Dec 1;208(11):1732-3. doi: 10.1093/infdis/jit509. Epub 2013 Sep 16. J Infect Dis. 2013. PMID: 24041794 No abstract available.
-
Risk of diarrhea during the first year of life associated with initial and subsequent colonization by specific enteropathogens.Am J Epidemiol. 1990 May;131(5):886-904. doi: 10.1093/oxfordjournals.aje.a115579. Am J Epidemiol. 1990. PMID: 2157338
-
Passive immunity against human pathogens using bovine antibodies.Clin Exp Immunol. 1999 May;116(2):193-205. doi: 10.1046/j.1365-2249.1999.00880.x. Clin Exp Immunol. 1999. PMID: 10337007 Free PMC article. Review. No abstract available.
Cited by
-
Antibiotic exposure among young infants suffering from diarrhoea in Bangladesh.J Paediatr Child Health. 2021 Mar;57(3):395-402. doi: 10.1111/jpc.15233. Epub 2020 Oct 27. J Paediatr Child Health. 2021. PMID: 33107165 Free PMC article.
-
Molecular characterization of diarrheagenic Escherichia coli pathotypes: Association of virulent genes, serogroups, and antibiotic resistance among moderate-to-severe diarrhea patients.J Clin Lab Anal. 2018 Jun;32(5):e22388. doi: 10.1002/jcla.22388. Epub 2018 Jan 21. J Clin Lab Anal. 2018. PMID: 29356079 Free PMC article.
-
Microbiota that affect risk for shigellosis in children in low-income countries.Emerg Infect Dis. 2015 Feb;21(2):242-50. doi: 10.3201/eid2101.140795. Emerg Infect Dis. 2015. PMID: 25625766 Free PMC article.
-
Child defecation and feces management practices in rural Bangladesh: Associations with fecal contamination, observed hand cleanliness and child diarrhea.PLoS One. 2020 Jul 20;15(7):e0236163. doi: 10.1371/journal.pone.0236163. eCollection 2020. PLoS One. 2020. PMID: 32687513 Free PMC article.
-
Impact of Microbiota: A Paradigm for Evolving Herd Immunity against Viral Diseases.Viruses. 2020 Oct 10;12(10):1150. doi: 10.3390/v12101150. Viruses. 2020. PMID: 33050511 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

