Arginine and disordered amyloid-β peptide structures: molecular level insights into the toxicity in Alzheimer's disease
- PMID: 24041422
- PMCID: PMC3867960
- DOI: 10.1021/cn4001389
Arginine and disordered amyloid-β peptide structures: molecular level insights into the toxicity in Alzheimer's disease
Abstract
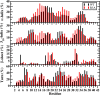
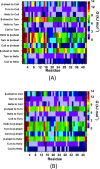
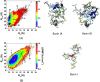
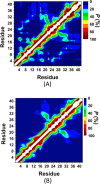
Recent studies present that the single arginine (R) residue in the sequence of Aβ42 adopts abundant β-sheet structure and forms stable salt bridges with various residues. Furthermore, experiments proposed that R stimulates the Aβ assembly and arginine (R) to alanine (A) mutation (R5A) decreases both aggregate formation tendency and the degree of its toxicity. However, the exact roles of R and R5A mutation in the structures of Aβ42 are poorly understood. Extensive molecular dynamics simulations along with thermodynamic calculations present that R5A mutation impacts the structures and free energy landscapes of the aqueous Aβ42 peptide. The β-sheet structure almost disappears in the Ala21-Ala30 region but is more abundant in parts of the central hydrophobic core and C-terminal regions of Aβ42 upon R5A mutation. More abundant α-helix is adopted in parts of the N-terminal and mid-domain regions and less prominent α-helix formation occurs in the central hydrophobic core region of Aβ42 upon R5A mutation. Interestingly, intramolecular interactions between N- and C-terminal or mid-domain regions disappear upon R5A mutation. The structures of Aβ42 are thermodynamically less stable and retain reduced compactness upon R5A mutation. R5A mutant-type structure stability increases with more prominent central hydrophobic core and mid-domain or C-terminal region interactions. Based on our results reported in this work, small organic molecules and antibodies that avoid β-sheet formation in the Ala21-Ala30 region and hinder the intramolecular interactions occurring between the N-terminal and mid-domain or C-terminal regions of Aβ42 may help to reduce Aβ42 toxicity in Alzheimer's disease.
Figures
Similar articles
-
The structures of the E22Δ mutant-type amyloid-β alloforms and the impact of E22Δ mutation on the structures of the wild-type amyloid-β alloforms.ACS Chem Neurosci. 2013 Feb 20;4(2):310-20. doi: 10.1021/cn300149j. Epub 2012 Dec 18. ACS Chem Neurosci. 2013. PMID: 23421682 Free PMC article.
-
Impact of K16A and K28A mutation on the structure and dynamics of amyloid-β42 peptide in Alzheimer's disease: key insights from molecular dynamics simulations.J Biomol Struct Dyn. 2020 Feb;38(3):708-721. doi: 10.1080/07391102.2019.1586587. Epub 2019 Apr 2. J Biomol Struct Dyn. 2020. PMID: 30821624
-
Structures and free energy landscapes of aqueous zinc(II)-bound amyloid-β(1-40) and zinc(II)-bound amyloid-β(1-42) with dynamics.J Biol Inorg Chem. 2012 Aug;17(6):927-38. doi: 10.1007/s00775-012-0909-9. Epub 2012 Jun 7. J Biol Inorg Chem. 2012. PMID: 22674434 Free PMC article.
-
Role of the region 23-28 in Abeta fibril formation: insights from simulations of the monomers and dimers of Alzheimer's peptides Abeta40 and Abeta42.Curr Alzheimer Res. 2008 Jun;5(3):244-50. doi: 10.2174/156720508784533330. Curr Alzheimer Res. 2008. PMID: 18537541 Review.
-
Understanding amyloid fibril nucleation and aβ oligomer/drug interactions from computer simulations.Acc Chem Res. 2014 Feb 18;47(2):603-11. doi: 10.1021/ar4002075. Epub 2013 Dec 24. Acc Chem Res. 2014. PMID: 24368046 Review.
Cited by
-
Divalent copper ion bound amyloid-β(40) and amyloid-β(42) alloforms are less preferred than divalent zinc ion bound amyloid-β(40) and amyloid-β(42) alloforms.J Biol Inorg Chem. 2016 Dec;21(8):957-973. doi: 10.1007/s00775-016-1392-5. Epub 2016 Sep 22. J Biol Inorg Chem. 2016. PMID: 27659954
-
Paving the Way for Synthetic Intrinsically Disordered Polymers for Soft Robotics.Polymers (Basel). 2023 Feb 2;15(3):763. doi: 10.3390/polym15030763. Polymers (Basel). 2023. PMID: 36772065 Free PMC article. Review.
-
How accurate are your simulations? Effects of confined aqueous volume and AMBER FF99SB and CHARMM22/CMAP force field parameters on structural ensembles of intrinsically disordered proteins: Amyloid-β42 in water.Intrinsically Disord Proteins. 2017 Oct 30;5(1):e1377813. doi: 10.1080/21690707.2017.1377813. eCollection 2017. Intrinsically Disord Proteins. 2017. PMID: 30250773 Free PMC article.
-
Structural Properties of Rat Intestinal Fatty Acid-Binding Protein with its Dynamics: Insights into Intrinsic Disorder.Protein Pept Lett. 2024;31(6):458-468. doi: 10.2174/0109298665313811240530055004. Protein Pept Lett. 2024. PMID: 38910419
-
Secondary structure dependence on simulation techniques and force field parameters: from disordered to ordered proteins.Biophys Rev. 2021 Oct 13;13(6):1173-1178. doi: 10.1007/s12551-021-00850-5. eCollection 2021 Dec. Biophys Rev. 2021. PMID: 35059035 Free PMC article. Review.
References
-
- Uversky V. N.; Oldfield C. J.; Dunker A. K. (2008) Intrinsically disordered proteins in human diseases: Introducing the D(2) concept. Annu. Rev. Biophys. 37, 215–246. - PubMed
-
- Babu M. M.; van der Lee R.; de Groot N. S.; Gsponer J. (2011) Intrinsically disordered proteins: Regulation and disease. Curr. Opin. Struct. Biol. 21, 432–440. - PubMed
-
- Dyson H. J.; Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208. - PubMed
-
- Wise-Scira O.; Xu L.; Kitahara T.; Perry G.; Coskuner O. (2011) Amyloid-β peptide structure in aqueous solution varies with fragment size. J. Chem. Phys. 135, 205101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

