An enhancer element harboring variants associated with systemic lupus erythematosus engages the TNFAIP3 promoter to influence A20 expression
- PMID: 24039598
- PMCID: PMC3764111
- DOI: 10.1371/journal.pgen.1003750
An enhancer element harboring variants associated with systemic lupus erythematosus engages the TNFAIP3 promoter to influence A20 expression
Abstract
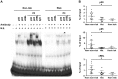
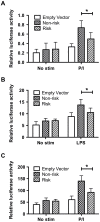
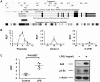
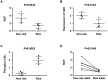
Functional characterization of causal variants present on risk haplotypes identified through genome-wide association studies (GWAS) is a primary objective of human genetics. In this report, we evaluate the function of a pair of tandem polymorphic dinucleotides, 42 kb downstream of the promoter of TNFAIP3, (rs148314165, rs200820567, collectively referred to as TT>A) recently nominated as causal variants responsible for genetic association of systemic lupus erythematosus (SLE) with tumor necrosis factor alpha inducible protein 3 (TNFAIP3). TNFAIP3 encodes the ubiquitin-editing enzyme, A20, a key negative regulator of NF-κB signaling. A20 expression is reduced in subjects carrying the TT>A risk alleles; however, the underlying functional mechanism by which this occurs is unclear. We used a combination of electrophoretic mobility shift assays (EMSA), mass spectrometry (MS), reporter assays, chromatin immunoprecipitation-PCR (ChIP-PCR) and chromosome conformation capture (3C) EBV transformed lymphoblastoid cell lines (LCL) from individuals carrying risk and non-risk TNFAIP3 haplotypes to characterize the effect of TT>A on A20 expression. Our results demonstrate that the TT>A variants reside in an enhancer element that binds NF-κB and SATB1 enabling physical interaction of the enhancer with the TNFAIP3 promoter through long-range DNA looping. Impaired binding of NF-κB to the TT>A risk alleles or knockdown of SATB1 expression by shRNA, inhibits the looping interaction resulting in reduced A20 expression. Together, these data reveal a novel mechanism of TNFAIP3 transcriptional regulation and establish the functional basis by which the TT>A risk variants attenuate A20 expression through inefficient delivery of NF-κB to the TNFAIP3 promoter. These results provide critical functional evidence supporting a direct causal role for TT>A in the genetic predisposition to SLE.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
TALEN-mediated enhancer knockout influences TNFAIP3 gene expression and mimics a molecular phenotype associated with systemic lupus erythematosus.Genes Immun. 2016 Apr;17(3):165-70. doi: 10.1038/gene.2016.4. Epub 2016 Jan 28. Genes Immun. 2016. PMID: 26821284 Free PMC article.
-
Functional variants of TNFAIP3 are associated with systemic lupus erythematosus in a cohort of Chinese Han population.Hum Immunol. 2019 Feb;80(2):140-145. doi: 10.1016/j.humimm.2018.11.008. Epub 2018 Dec 5. Hum Immunol. 2019. PMID: 30529365
-
Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus.Nat Genet. 2011 Mar;43(3):253-8. doi: 10.1038/ng.766. Epub 2011 Feb 20. Nat Genet. 2011. PMID: 21336280 Free PMC article.
-
Single nucleotide polymorphisms at the TNFAIP3/A20 locus and susceptibility/resistance to inflammatory and autoimmune diseases.Adv Exp Med Biol. 2014;809:163-83. doi: 10.1007/978-1-4939-0398-6_10. Adv Exp Med Biol. 2014. PMID: 25302371 Review.
-
The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology.Trends Immunol. 2009 Aug;30(8):383-91. doi: 10.1016/j.it.2009.05.007. Epub 2009 Jul 28. Trends Immunol. 2009. PMID: 19643665 Review.
Cited by
-
A20 at the Crossroads of Cell Death, Inflammation, and Autoimmunity.Cold Spring Harb Perspect Biol. 2020 Jan 2;12(1):a036418. doi: 10.1101/cshperspect.a036418. Cold Spring Harb Perspect Biol. 2020. PMID: 31427375 Free PMC article. Review.
-
Genetics of systemic lupus erythematosus: immune responses and end organ resistance to damage.Curr Opin Immunol. 2014 Dec;31:87-96. doi: 10.1016/j.coi.2014.10.004. Epub 2014 Oct 25. Curr Opin Immunol. 2014. PMID: 25458999 Free PMC article. Review.
-
TALEN-mediated enhancer knockout influences TNFAIP3 gene expression and mimics a molecular phenotype associated with systemic lupus erythematosus.Genes Immun. 2016 Apr;17(3):165-70. doi: 10.1038/gene.2016.4. Epub 2016 Jan 28. Genes Immun. 2016. PMID: 26821284 Free PMC article.
-
Low TNFAIP3 expression in psoriatic skin promotes disease susceptibility and severity.PLoS One. 2019 May 23;14(5):e0217352. doi: 10.1371/journal.pone.0217352. eCollection 2019. PLoS One. 2019. PMID: 31120955 Free PMC article.
-
The New Frontier of Functional Genomics: From Chromatin Architecture and Noncoding RNAs to Therapeutic Targets.SLAS Discov. 2020 Jul;25(6):568-580. doi: 10.1177/2472555220926158. Epub 2020 Jun 2. SLAS Discov. 2020. PMID: 32486876 Free PMC article.
References
-
- Jaattela M, Mouritzen H, Elling F, Bastholm L (1996) A20 zinc finger protein inhibits TNF and IL-1 signaling. J Immunol 156: 1166–1173. - PubMed
-
- Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, et al. (2004) The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol 5: 1052–1060. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

