The plant host can affect the encapsidation of brome mosaic virus (BMV) RNA: BMV virions are surprisingly heterogeneous
- PMID: 24036424
- PMCID: PMC3944473
- DOI: 10.1016/j.jmb.2013.09.007
The plant host can affect the encapsidation of brome mosaic virus (BMV) RNA: BMV virions are surprisingly heterogeneous
Abstract
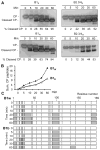
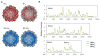
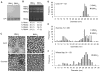
Brome mosaic virus (BMV) packages its genomic and subgenomic RNAs into three separate viral particles. BMV purified from barley, wheat, and tobacco have distinct relative abundances of the encapsidated RNAs. We seek to identify the basis for the host-dependent differences in viral RNA encapsidation. Sequencing of the viral RNAs revealed recombination events in the 3' untranslated region of RNA1 of BMV purified from barley and wheat, but not from tobacco. However, the relative amounts of the BMV RNAs that accumulated in barley and wheat are similar and RNA accumulation is not sufficient to account for the difference in RNA encapsidation. Virions purified from barley and wheat were found to differ in their isoelectric points, resistance to proteolysis, and contacts between the capsid residues and the RNA. Mass spectrometric analyses revealed that virions from the three hosts had different post-translational modifications that should impact the physiochemical properties of the virions. Another major source of variation in RNA encapsidation was due to the purification of BMV particles to homogeneity. Highly enriched BMV present in lysates had a surprising range of sizes, buoyant densities, and distinct relative amounts of encapsidated RNAs. These results show that the encapsidated BMV RNAs reflect a combination of host effects on the physiochemical properties of the viral capsids and the enrichment of a subset of virions. The previously unexpected heterogeneity in BMV should influence the timing of the infection and also the host innate immune responses.
Keywords: Brome mosaic virus; RNA encapsidation; capsid–RNA interaction; coat protein; virion polymorphism.
© 2013.
Figures





Similar articles
-
Phosphorylation of the Brome Mosaic Virus Capsid Regulates the Timing of Viral Infection.J Virol. 2016 Aug 12;90(17):7748-60. doi: 10.1128/JVI.00833-16. Print 2016 Sep 1. J Virol. 2016. PMID: 27334588 Free PMC article.
-
Unravelling the Stability and Capsid Dynamics of the Three Virions of Brome Mosaic Virus Assembled Autonomously In Vivo.J Virol. 2020 Mar 31;94(8):e01794-19. doi: 10.1128/JVI.01794-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31996436 Free PMC article.
-
The tripartite virions of the brome mosaic virus have distinct physical properties that affect the timing of the infection process.J Virol. 2014 Jun;88(11):6483-91. doi: 10.1128/JVI.00377-14. Epub 2014 Mar 26. J Virol. 2014. PMID: 24672042 Free PMC article.
-
The coat protein leads the way: an update on basic and applied studies with the Brome mosaic virus coat protein.Mol Plant Pathol. 2011 May;12(4):403-12. doi: 10.1111/j.1364-3703.2010.00678.x. Epub 2010 Nov 25. Mol Plant Pathol. 2011. PMID: 21453435 Free PMC article. Review.
-
The brome mosaic virus 3' untranslated sequence regulates RNA replication, recombination, and virion assembly.Virus Res. 2015 Aug 3;206:46-52. doi: 10.1016/j.virusres.2015.02.007. Epub 2015 Feb 14. Virus Res. 2015. PMID: 25687214 Review.
Cited by
-
Bacteriophage MS2 genomic RNA encodes an assembly instruction manual for its capsid.Bacteriophage. 2016 Mar 2;6(1):e1157666. doi: 10.1080/21597081.2016.1157666. eCollection 2016 Jan-Mar. Bacteriophage. 2016. PMID: 27144089 Free PMC article.
-
Within-host Evolution of Segments Ratio for the Tripartite Genome of Alfalfa Mosaic Virus.Sci Rep. 2017 Jul 10;7(1):5004. doi: 10.1038/s41598-017-05335-8. Sci Rep. 2017. PMID: 28694514 Free PMC article.
-
Mapping RNA Sequences that Contact Viral Capsid Proteins in Virions.Bio Protoc. 2014 Jul 20;7(14):e2398. doi: 10.21769/BioProtoc.2398. Bio Protoc. 2014. PMID: 29082288 Free PMC article.
-
Compositional complementarity between genomic RNA and coat proteins in positive-sense single-stranded RNA viruses.Nucleic Acids Res. 2022 Apr 22;50(7):4054-4067. doi: 10.1093/nar/gkac202. Nucleic Acids Res. 2022. PMID: 35357492 Free PMC article.
-
Modulation of Capsid Dynamics in Bromoviruses by the Host and Heterologous Viral Replicase.J Virol. 2023 Mar 30;97(3):e0128422. doi: 10.1128/jvi.01284-22. Epub 2023 Feb 14. J Virol. 2023. PMID: 36786601 Free PMC article.
References
-
- Levine B, Hardwick JM, Griffin DE. Persistence of alphaviruses in vertebrate hosts. Trends Microbiol. 1994;2:25–28. - PubMed
-
- Rao AL. Genome packaging by spherical plant RNA viruses. Annu Rev Phytopathol. 2006;44:61–87. - PubMed
-
- Lane LC. The bromoviruses. Adv Virus Res. 1974;19:151–220. - PubMed
-
- Rao A, Grantham G. Biological significance of the seven amino-terminal basic residues of brome mosaic virus coat protein. Virology. 1995;211:42–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

