Uncoupling of allosteric and oligomeric regulation in a functional hybrid enzyme constructed from Escherichia coli and human ribonucleotide reductase
- PMID: 24024562
- PMCID: PMC8382989
- DOI: 10.1021/bi400781z
Uncoupling of allosteric and oligomeric regulation in a functional hybrid enzyme constructed from Escherichia coli and human ribonucleotide reductase
Abstract
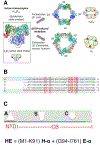
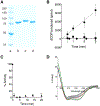
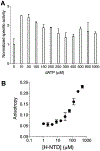
An N-terminal-domain (NTD) and adjacent catalytic body (CB) make up subunit-α of ribonucleotide reductase (RNR), the rate-limiting enzyme for de novo dNTP biosynthesis. A strong linkage exists between ligand binding at the NTD and oligomerization-coupled RNR inhibition, inducible by both dATP and nucleotide chemotherapeutics. These observations have distinguished the NTD as an oligomeric regulation domain dictating the assembly of inactive RNR oligomers. Inactive states of RNR differ between eukaryotes and prokaryotes (α6 in human versus α4β4 in Escherichia coli , wherein β is RNR's other subunit); however, the NTD structurally interconnects individual α2 or α2 and β2 dimeric motifs within the respective α6 or α4β4 complexes. To elucidate the influence of NTD ligand binding on RNR allosteric and oligomeric regulation, we engineered a human- E. coli hybrid enzyme (HE) where human-NTD is fused to E. coli -CB. Both the NTD and the CB of the HE bind dATP. The HE specifically partners with E. coli -β to form an active holocomplex. However, although the NTD is the sole physical tether to support α2 and/or β2 associations in the dATP-bound α6 or α4β4 fully inhibited RNR complexes, the binding of dATP to the HE NTD only partially suppresses HE activity and fully precludes formation of higher-order HE oligomers. We postulate that oligomeric regulation is the ultimate mechanism for potent RNR inhibition, requiring species-specific NTD-CB interactions. Such interdomain cooperativity in RNR oligomerization is unexpected from structural studies alone or biochemical studies of point mutants.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Disruption of an oligomeric interface prevents allosteric inhibition of Escherichia coli class Ia ribonucleotide reductase.J Biol Chem. 2018 Jun 29;293(26):10404-10412. doi: 10.1074/jbc.RA118.002569. Epub 2018 Apr 26. J Biol Chem. 2018. PMID: 29700111 Free PMC article.
-
Diversity in Overall Activity Regulation of Ribonucleotide Reductase.J Biol Chem. 2015 Jul 10;290(28):17339-48. doi: 10.1074/jbc.M115.649624. Epub 2015 May 13. J Biol Chem. 2015. PMID: 25971975 Free PMC article.
-
Basis of dATP inhibition of RNRs.J Biol Chem. 2018 Jun 29;293(26):10413-10414. doi: 10.1074/jbc.H118.003717. J Biol Chem. 2018. PMID: 29959279 Free PMC article.
-
DNA building blocks: keeping control of manufacture.Crit Rev Biochem Mol Biol. 2012 Jan-Feb;47(1):50-63. doi: 10.3109/10409238.2011.630372. Epub 2011 Nov 3. Crit Rev Biochem Mol Biol. 2012. PMID: 22050358 Free PMC article. Review.
-
The prototypic class Ia ribonucleotide reductase from Escherichia coli: still surprising after all these years.Biochem Soc Trans. 2012 Jun 1;40(3):523-30. doi: 10.1042/BST20120081. Biochem Soc Trans. 2012. PMID: 22616862 Free PMC article. Review.
Cited by
-
The more the merrier: how homo-oligomerization alters the interactome and function of ribonucleotide reductase.Curr Opin Chem Biol. 2020 Feb;54:10-18. doi: 10.1016/j.cbpa.2019.09.003. Epub 2019 Nov 15. Curr Opin Chem Biol. 2020. PMID: 31734537 Free PMC article. Review.
-
Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies.Oncogene. 2015 Apr 16;34(16):2011-21. doi: 10.1038/onc.2014.155. Epub 2014 Jun 9. Oncogene. 2015. PMID: 24909171 Review.
-
Cladribine and Fludarabine Nucleotides Induce Distinct Hexamers Defining a Common Mode of Reversible RNR Inhibition.ACS Chem Biol. 2016 Jul 15;11(7):2021-32. doi: 10.1021/acschembio.6b00303. Epub 2016 May 20. ACS Chem Biol. 2016. PMID: 27159113 Free PMC article.
-
Still no Rest for the Reductases: Ribonucleotide Reductase (RNR) Structure and Function: An Update.Subcell Biochem. 2022;99:155-197. doi: 10.1007/978-3-031-00793-4_5. Subcell Biochem. 2022. PMID: 36151376 Review.
-
Clofarabine Commandeers the RNR-α-ZRANB3 Nuclear Signaling Axis.Cell Chem Biol. 2020 Jan 16;27(1):122-133.e5. doi: 10.1016/j.chembiol.2019.11.012. Epub 2019 Dec 10. Cell Chem Biol. 2020. PMID: 31836351 Free PMC article.
References
-
- Fersht A (1999) Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding, W. H. Freeman, New York.
-
- Walsh C (1979) Enzymatic reaction mechanisms, W. H. Freeman, San Francisco.
-
- Hedstrom L (2002) Serine protease mechanism and specificity. Chem. Rev. (Washington, DC, U. S.) 102, 4501–4524. - PubMed
-
- Britt BM (1997) For enzymes, bigger is better. Biophys. Chem 69, 63–70. - PubMed
-
- Srere PA (1984) Why are enzymes so big? Trends Biochem. Sci 9, 387–390.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

