Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance
- PMID: 24012370
- PMCID: PMC3934494
- DOI: 10.1016/j.chembiol.2013.07.014
Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance
Abstract
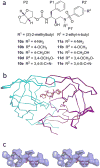
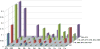
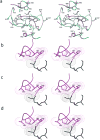
The rapid evolution of HIV under selective drug pressure has led to multidrug resistant (MDR) strains that evade standard therapies. We designed highly potent HIV-1 protease inhibitors (PIs) using the substrate envelope model, which confines inhibitors within the consensus volume of natural substrates, providing inhibitors less susceptible to resistance because a mutation affecting such inhibitors will simultaneously affect viral substrate processing. The designed PIs share a common chemical scaffold but utilize various moieties that optimally fill the substrate envelope, as confirmed by crystal structures. The designed PIs retain robust binding to MDR protease variants and display exceptional antiviral potencies against different clades of HIV as well as a panel of 12 drug-resistant viral strains. The substrate envelope model proves to be a powerful strategy to develop potent and robust inhibitors that avoid drug resistance.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
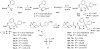
Similar articles
-
Design, synthesis, and biological and structural evaluations of novel HIV-1 protease inhibitors to combat drug resistance.J Med Chem. 2012 Jul 26;55(14):6328-41. doi: 10.1021/jm300238h. Epub 2012 Jul 13. J Med Chem. 2012. PMID: 22708897 Free PMC article.
-
Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance.J Virol. 2010 May;84(10):5368-78. doi: 10.1128/JVI.02531-09. Epub 2010 Mar 17. J Virol. 2010. PMID: 20237088 Free PMC article.
-
HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.J Am Chem Soc. 2008 May 14;130(19):6099-113. doi: 10.1021/ja076558p. Epub 2008 Apr 16. J Am Chem Soc. 2008. PMID: 18412349 Free PMC article.
-
Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir.AIDS Rev. 2008 Jul-Sep;10(3):131-42. AIDS Rev. 2008. PMID: 18820715 Free PMC article. Review.
-
Design of HIV protease inhibitors targeting protein backbone: an effective strategy for combating drug resistance.Acc Chem Res. 2008 Jan;41(1):78-86. doi: 10.1021/ar7001232. Epub 2007 Aug 28. Acc Chem Res. 2008. PMID: 17722874 Review.
Cited by
-
Interdependence of Inhibitor Recognition in HIV-1 Protease.J Chem Theory Comput. 2017 May 9;13(5):2300-2309. doi: 10.1021/acs.jctc.6b01262. Epub 2017 Apr 11. J Chem Theory Comput. 2017. PMID: 28358514 Free PMC article.
-
HIV-1 Integrase Inhibitors with Modifications That Affect Their Potencies against Drug Resistant Integrase Mutants.ACS Infect Dis. 2021 Jun 11;7(6):1469-1482. doi: 10.1021/acsinfecdis.0c00819. Epub 2021 Mar 9. ACS Infect Dis. 2021. PMID: 33686850 Free PMC article.
-
Molecular and Dynamic Mechanism Underlying Drug Resistance in Genotype 3 Hepatitis C NS3/4A Protease.J Am Chem Soc. 2016 Sep 14;138(36):11850-9. doi: 10.1021/jacs.6b06454. Epub 2016 Sep 2. J Am Chem Soc. 2016. PMID: 27512818 Free PMC article.
-
An NMR strategy to detect conformational differences in a protein complexed with highly analogous inhibitors in solution.Methods. 2018 Sep 15;148:9-18. doi: 10.1016/j.ymeth.2018.04.005. Epub 2018 Apr 12. Methods. 2018. PMID: 29656080 Free PMC article.
-
CRISPR-Cas9-mediated saturated mutagenesis screen predicts clinical drug resistance with improved accuracy.Proc Natl Acad Sci U S A. 2017 Oct 31;114(44):11751-11756. doi: 10.1073/pnas.1708268114. Epub 2017 Oct 16. Proc Natl Acad Sci U S A. 2017. PMID: 29078326 Free PMC article.
References
-
- Collaborative Computational Project Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. - PubMed
-
- Stanford HIV Drug Resistance Database. [accessed on 02 July, 2013]; http://hivdb.Stanford.edu.
-
- U.S. Food and Drug Administration. [accessed on 02 July, 2013];Antiretroviral drugs used in the treatment of HIV infection. http://www.fda.gov/ForConsumers/ByAudience/ForPatientAdvocates/HIVandAID....
-
- Ali A, Reddy GSKK, Cao H, Anjum SG, Nalam MNL, Schiffer CA, Rana TM. Discovery of HIV-1 protease inhibitors with picomolar affinities incorporating N-aryl-oxazolidinone-5-carboxamides as novel P2 ligands. J Med Chem. 2006;49:7342–7356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 GM103543/GM/NIGMS NIH HHS/United States
- AI41404/AI/NIAID NIH HHS/United States
- R01 AI043198/AI/NIAID NIH HHS/United States
- P41 RR007707/RR/NCRR NIH HHS/United States
- P01-GM66524/GM/NIGMS NIH HHS/United States
- R01 AI041404/AI/NIAID NIH HHS/United States
- R01 GM082209/GM/NIGMS NIH HHS/United States
- GM065418/GM/NIGMS NIH HHS/United States
- S10 OD012028/OD/NIH HHS/United States
- R01 DA030199/DA/NIDA NIH HHS/United States
- GM082209/GM/NIGMS NIH HHS/United States
- P01GM066524-08S1/GM/NIGMS NIH HHS/United States
- P01 MH100942/MH/NIMH NIH HHS/United States
- RR007707/RR/NCRR NIH HHS/United States
- AI43198/AI/NIAID NIH HHS/United States
- P01 GM066524/GM/NIGMS NIH HHS/United States
- R24 GM111072/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

