Small RNAs derived from lncRNA RNase MRP have gene-silencing activity relevant to human cartilage-hair hypoplasia
- PMID: 24009312
- PMCID: PMC3869355
- DOI: 10.1093/hmg/ddt427
Small RNAs derived from lncRNA RNase MRP have gene-silencing activity relevant to human cartilage-hair hypoplasia
Abstract
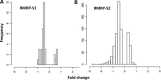
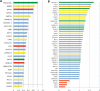
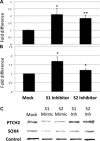
Post-transcriptional processing of some long non-coding RNAs (lncRNAs) reveals that they are a source of miRNAs. We show that the 268-nt non-coding RNA component of mitochondrial RNA processing endoribonuclease, (RNase MRP), is the source of at least two short (∼20 nt) RNAs designated RMRP-S1 and RMRP-S2, which function as miRNAs. Point mutations in RNase MRP cause human cartilage-hair hypoplasia (CHH), and several disease-causing mutations map to RMRP-S1 and -S2. SHAPE chemical probing identified two alternative secondary structures altered by disease mutations. RMRP-S1 and -S2 are significantly reduced in two fibroblast cell lines and a B-cell line derived from CHH patients. Tests of gene regulatory activity of RMRP-S1 and -S2 identified over 900 genes that were significantly regulated, of which over 75% were down-regulated, and 90% contained target sites with seed complements of RMRP-S1 and -S2 predominantly in their 3' UTRs. Pathway analysis identified regulated genes that function in skeletal development, hair development and hematopoietic cell differentiation including PTCH2 and SOX4 among others, linked to major CHH phenotypes. Also, genes associated with alternative RNA splicing, cell proliferation and differentiation were highly targeted. Therefore, alterations RMRP-S1 and -S2, caused by point mutations in RMRP, are strongly implicated in the molecular mechanism of CHH.
Figures



Similar articles
-
Expression of RMRP RNA is regulated in chondrocyte hypertrophy and determines chondrogenic differentiation.Sci Rep. 2017 Jul 25;7(1):6440. doi: 10.1038/s41598-017-06809-5. Sci Rep. 2017. PMID: 28743979 Free PMC article.
-
Defects in lymphocyte telomere homeostasis contribute to cellular immune phenotype in patients with cartilage-hair hypoplasia.J Allergy Clin Immunol. 2017 Oct;140(4):1120-1129.e1. doi: 10.1016/j.jaci.2016.11.051. Epub 2017 Jan 23. J Allergy Clin Immunol. 2017. PMID: 28126377
-
A disease-linked lncRNA mutation in RNase MRP inhibits ribosome synthesis.Nat Commun. 2022 Feb 3;13(1):649. doi: 10.1038/s41467-022-28295-8. Nat Commun. 2022. PMID: 35115551 Free PMC article.
-
RNase MRP and disease.Wiley Interdiscip Rev RNA. 2010 Jul-Aug;1(1):102-16. doi: 10.1002/wrna.9. Epub 2010 May 6. Wiley Interdiscip Rev RNA. 2010. PMID: 21956908 Review.
-
RMRP-related short stature: A report of six additional Japanese individuals with cartilage hair hypoplasia and literature review.Am J Med Genet A. 2024 Jun;194(6):e63562. doi: 10.1002/ajmg.a.63562. Epub 2024 Feb 9. Am J Med Genet A. 2024. PMID: 38337186 Review.
Cited by
-
The role of long non-coding RNAs in the pathogenesis of hereditary diseases.BMC Med Genomics. 2019 Mar 13;12(Suppl 2):42. doi: 10.1186/s12920-019-0487-6. BMC Med Genomics. 2019. PMID: 30871545 Free PMC article. Review.
-
LncRNA RMRP/miR-613 axis is associated with poor prognosis and enhances the tumorigenesis of hepatocellular carcinoma by impacting oncogenic phenotypes.Am J Transl Res. 2019 May 15;11(5):2801-2815. eCollection 2019. Am J Transl Res. 2019. PMID: 31217855 Free PMC article.
-
The roles of RNA processing in translating genotype to phenotype.Nat Rev Mol Cell Biol. 2017 Feb;18(2):102-114. doi: 10.1038/nrm.2016.139. Epub 2016 Nov 16. Nat Rev Mol Cell Biol. 2017. PMID: 27847391 Free PMC article. Review.
-
Ribosomopathies: Global process, tissue specific defects.Rare Dis. 2015 Apr 1;3(1):e1025185. doi: 10.1080/21675511.2015.1025185. eCollection 2015. Rare Dis. 2015. PMID: 26442198 Free PMC article. Review.
-
Non-coding RNAs and disease: the classical ncRNAs make a comeback.Biochem Soc Trans. 2016 Aug 15;44(4):1073-8. doi: 10.1042/BST20160089. Biochem Soc Trans. 2016. PMID: 27528754 Free PMC article. Review.
References
-
- Ridanpaa M., van Eenennaam H., Pelin K., Chadwick R., Johnson C., Yuan B., van Venrooij W., Pruijn G., Salmela R., Rockas S., et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. - PubMed
-
- Ridanpaa M., Jain P., McKusick V.A., Francomano C.A., Kaitila I. The major mutation in the RMRP gene causing CHH among the Amish is the same as that found in most Finnish cases. Am. J. Med. Genet. 2003;121C:81–83. - PubMed
-
- Hermanns P., Tran A., Munivez E., Carter S., Zabel B., Lee B., Leroy J.G. RMRP Mutations in cartilage-hair hypoplasia. Am. J. Med. Genet. 2006;140:2121–2130. - PubMed
-
- McKusick V.A., Eldridge R., Hostetler J.A., Ruangwit U., Egeland J.A. Dwarfism in the Amish. Ii. Cartilage-Hair Hypoplasia. Bull. Johns Hopkins Hosp. 1965;116:285–326. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

