Infernal 1.1: 100-fold faster RNA homology searches
- PMID: 24008419
- PMCID: PMC3810854
- DOI: 10.1093/bioinformatics/btt509
Infernal 1.1: 100-fold faster RNA homology searches
Abstract
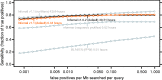
Summary: Infernal builds probabilistic profiles of the sequence and secondary structure of an RNA family called covariance models (CMs) from structurally annotated multiple sequence alignments given as input. Infernal uses CMs to search for new family members in sequence databases and to create potentially large multiple sequence alignments. Version 1.1 of Infernal introduces a new filter pipeline for RNA homology search based on accelerated profile hidden Markov model (HMM) methods and HMM-banded CM alignment methods. This enables ∼100-fold acceleration over the previous version and ∼10 000-fold acceleration over exhaustive non-filtered CM searches.
Availability: Source code, documentation and the benchmark are downloadable from http://infernal.janelia.org. Infernal is freely licensed under the GNU GPLv3 and should be portable to any POSIX-compliant operating system, including Linux and Mac OS/X. Documentation includes a user's guide with a tutorial, a discussion of file formats and user options and additional details on methods implemented in the software.
Contact: nawrockie@janelia.hhmi.org
Figures
Similar articles
-
Infernal 1.0: inference of RNA alignments.Bioinformatics. 2009 May 15;25(10):1335-7. doi: 10.1093/bioinformatics/btp157. Epub 2009 Mar 23. Bioinformatics. 2009. PMID: 19307242 Free PMC article.
-
nhmmer: DNA homology search with profile HMMs.Bioinformatics. 2013 Oct 1;29(19):2487-9. doi: 10.1093/bioinformatics/btt403. Epub 2013 Jul 9. Bioinformatics. 2013. PMID: 23842809 Free PMC article.
-
Query-dependent banding (QDB) for faster RNA similarity searches.PLoS Comput Biol. 2007 Mar 30;3(3):e56. doi: 10.1371/journal.pcbi.0030056. Epub 2007 Feb 7. PLoS Comput Biol. 2007. PMID: 17397253 Free PMC article.
-
Computational identification of functional RNA homologs in metagenomic data.RNA Biol. 2013 Jul;10(7):1170-9. doi: 10.4161/rna.25038. Epub 2013 May 20. RNA Biol. 2013. PMID: 23722291 Free PMC article. Review.
-
The art of editing RNA structural alignments.Methods Mol Biol. 2014;1097:379-94. doi: 10.1007/978-1-62703-709-9_17. Methods Mol Biol. 2014. PMID: 24639168 Review.
Cited by
-
DecoyFinder: Identification of Contaminants in Sets of Homologous RNA Sequences.bioRxiv [Preprint]. 2024 Oct 15:2024.10.12.618037. doi: 10.1101/2024.10.12.618037. bioRxiv. 2024. PMID: 39464058 Free PMC article. Preprint.
-
DRAGoM: Classification and Quantification of Noncoding RNA in Metagenomic Data.Front Genet. 2021 May 5;12:669495. doi: 10.3389/fgene.2021.669495. eCollection 2021. Front Genet. 2021. PMID: 34025724 Free PMC article.
-
Analysis of the Coptis chinensis genome reveals the diversification of protoberberine-type alkaloids.Nat Commun. 2021 Jun 2;12(1):3276. doi: 10.1038/s41467-021-23611-0. Nat Commun. 2021. PMID: 34078898 Free PMC article.
-
Insights into the Mechanism of Pre-mRNA Splicing of Tiny Introns from the Genome of a Giant Ciliate Stentor coeruleus.Int J Mol Sci. 2022 Sep 19;23(18):10973. doi: 10.3390/ijms231810973. Int J Mol Sci. 2022. PMID: 36142882 Free PMC article.
-
Chromosome-level genome assembly of cotton thrips Thrips tabaci (Thysanoptera: Thripidae).Sci Data. 2024 Sep 16;11(1):1003. doi: 10.1038/s41597-024-03737-8. Sci Data. 2024. PMID: 39294155 Free PMC article.
References
-
- Brown MP. Small subunit ribosomal RNA modeling using stochastic context-free grammars. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2000;8:57–66. - PubMed
-
- Durbin R, et al. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge, UK: Cambridge University Press; 1998.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

