Identification and characterization of multiple novel Rab-myosin Va interactions
- PMID: 24006491
- PMCID: PMC3814135
- DOI: 10.1091/mbc.E13-05-0236
Identification and characterization of multiple novel Rab-myosin Va interactions
Abstract
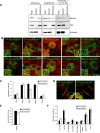
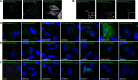
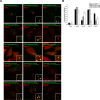
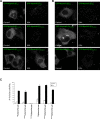
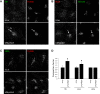
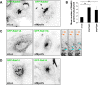
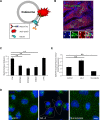
Myosin Va is a widely expressed actin-based motor protein that binds members of the Rab GTPase family (3A, 8A, 10, 11A, 27A) and is implicated in many intracellular trafficking processes. To our knowledge, myosin Va has not been tested in a systematic screen for interactions with the entire Rab GTPase family. To that end, we report a yeast two-hybrid screen of all human Rabs for myosin Va-binding ability and reveal 10 novel interactions (3B, 3C, 3D, 6A, 6A', 6B, 11B, 14, 25, 39B), which include interactions with three new Rab subfamilies (Rab6, Rab14, Rab39B). Of interest, myosin Va interacts with only a subset of the Rabs associated with the endocytic recycling and post-Golgi secretory systems. We demonstrate that myosin Va has three distinct Rab-binding domains on disparate regions of the motor (central stalk, an alternatively spliced exon, and the globular tail). Although the total pool of myosin Va is shared by several Rabs, Rab10 and Rab11 appear to be the major determinants of its recruitment to intracellular membranes. We also present evidence that myosin Va is necessary for maintaining a peripheral distribution of Rab11- and Rab14-positive endosomes.
Figures

Similar articles
-
LRRK2-phosphorylated Rab10 sequesters Myosin Va with RILPL2 during ciliogenesis blockade.Life Sci Alliance. 2021 Mar 16;4(5):e202101050. doi: 10.26508/lsa.202101050. Print 2021 May. Life Sci Alliance. 2021. PMID: 33727250 Free PMC article.
-
Ypt32p and Mlc1p bind within the vesicle binding region of the class V myosin Myo2p globular tail domain.Mol Microbiol. 2008 Mar;67(5):1051-66. doi: 10.1111/j.1365-2958.2008.06106.x. Epub 2008 Jan 23. Mol Microbiol. 2008. PMID: 18221262
-
In vitro reconstitution of a transport complex containing Rab27a, melanophilin and myosin Va.FEBS Lett. 2006 Oct 30;580(25):5863-8. doi: 10.1016/j.febslet.2006.09.047. Epub 2006 Oct 2. FEBS Lett. 2006. PMID: 17045265
-
Rab GTPases and myosin motors in organelle motility.Traffic. 2004 Jun;5(6):393-9. doi: 10.1111/j.1398-9219.2004.00190.x. Traffic. 2004. PMID: 15117313 Review.
-
Griscelli syndrome: a model system to study vesicular trafficking.Pigment Cell Melanoma Res. 2009 Jun;22(3):268-82. doi: 10.1111/j.1755-148X.2009.00558.x. Epub 2009 Feb 25. Pigment Cell Melanoma Res. 2009. PMID: 19243575 Review.
Cited by
-
Coronavirus M Protein Trafficking in Epithelial Cells Utilizes a Myosin Vb Splice Variant and Rab10.Cells. 2024 Jan 10;13(2):126. doi: 10.3390/cells13020126. Cells. 2024. PMID: 38247817 Free PMC article.
-
Roles and regulation of myosin V interaction with cargo.Adv Biol Regul. 2021 Jan;79:100787. doi: 10.1016/j.jbior.2021.100787. Epub 2021 Jan 20. Adv Biol Regul. 2021. PMID: 33541831 Free PMC article. Review.
-
F-actin disassembly factor MICAL1 binding to Myosin Va mediates cargo unloading during cytokinesis.Sci Adv. 2020 Nov 6;6(45):eabb1307. doi: 10.1126/sciadv.abb1307. Print 2020 Nov. Sci Adv. 2020. PMID: 33158857 Free PMC article.
-
Specific Light-Up System for Protein and Metabolite Targets Triggered by Initiation Complex Formation.Sci Rep. 2017 Nov 9;7(1):15191. doi: 10.1038/s41598-017-15697-8. Sci Rep. 2017. PMID: 29123195 Free PMC article.
-
LRRK2-phosphorylated Rab10 sequesters Myosin Va with RILPL2 during ciliogenesis blockade.Life Sci Alliance. 2021 Mar 16;4(5):e202101050. doi: 10.26508/lsa.202101050. Print 2021 May. Life Sci Alliance. 2021. PMID: 33727250 Free PMC article.
References
-
- Darchen F, Goud B. Multiple aspects of Rab protein action in the secretory pathway: focus on Rab3 and Rab6. Biochimie. 2000;82:375–384. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

