An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules
- PMID: 23993102
- PMCID: PMC3954635
- DOI: 10.1016/j.cell.2013.08.003
An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules
Abstract
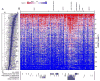
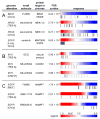
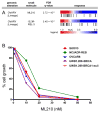
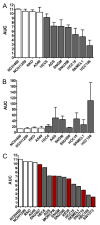
The high rate of clinical response to protein-kinase-targeting drugs matched to cancer patients with specific genomic alterations has prompted efforts to use cancer cell line (CCL) profiling to identify additional biomarkers of small-molecule sensitivities. We have quantitatively measured the sensitivity of 242 genomically characterized CCLs to an Informer Set of 354 small molecules that target many nodes in cell circuitry, uncovering protein dependencies that: (1) associate with specific cancer-genomic alterations and (2) can be targeted by small molecules. We have created the Cancer Therapeutics Response Portal (http://www.broadinstitute.org/ctrp) to enable users to correlate genetic features to sensitivity in individual lineages and control for confounding factors of CCL profiling. We report a candidate dependency, associating activating mutations in the oncogene β-catenin with sensitivity to the Bcl-2 family antagonist, navitoclax. The resource can be used to develop novel therapeutic hypotheses and to accelerate discovery of drugs matched to patients by their cancer genotype and lineage.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Harnessing Connectivity in a Large-Scale Small-Molecule Sensitivity Dataset.Cancer Discov. 2015 Nov;5(11):1210-23. doi: 10.1158/2159-8290.CD-15-0235. Epub 2015 Oct 19. Cancer Discov. 2015. PMID: 26482930 Free PMC article.
-
More than fishing for a cure: The promises and pitfalls of high throughput cancer cell line screens.Pharmacol Ther. 2018 Nov;191:178-189. doi: 10.1016/j.pharmthera.2018.06.014. Epub 2018 Jun 25. Pharmacol Ther. 2018. PMID: 29953899 Free PMC article. Review.
-
Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells.Nucleic Acids Res. 2013 Jan;41(Database issue):D955-61. doi: 10.1093/nar/gks1111. Epub 2012 Nov 23. Nucleic Acids Res. 2013. PMID: 23180760 Free PMC article.
-
A Road Map to Personalizing Targeted Cancer Therapies Using Synthetic Lethality.Trends Cancer. 2019 Jan;5(1):11-29. doi: 10.1016/j.trecan.2018.11.001. Epub 2018 Dec 7. Trends Cancer. 2019. PMID: 30616753 Review.
-
MDP, a database linking drug response data to genomic information, identifies dasatinib and statins as a combinatorial strategy to inhibit YAP/TAZ in cancer cells.Oncotarget. 2015 Nov 17;6(36):38854-65. doi: 10.18632/oncotarget.5749. Oncotarget. 2015. PMID: 26513174 Free PMC article.
Cited by
-
Current status and perspectives of patient-derived rare cancer models.Hum Cell. 2020 Oct;33(4):919-929. doi: 10.1007/s13577-020-00391-1. Epub 2020 Jun 14. Hum Cell. 2020. PMID: 32537685 Review.
-
Using Pharmacogenomic Databases for Discovering Patient-Target Genes and Small Molecule Candidates to Cancer Therapy.Front Pharmacol. 2016 Sep 29;7:312. doi: 10.3389/fphar.2016.00312. eCollection 2016. Front Pharmacol. 2016. PMID: 27746730 Free PMC article. Review.
-
Novel carfilzomib-based combinations as potential therapeutic strategies for liposarcomas.Cell Mol Life Sci. 2021 Feb;78(4):1837-1851. doi: 10.1007/s00018-020-03620-w. Epub 2020 Aug 26. Cell Mol Life Sci. 2021. PMID: 32851475 Free PMC article.
-
Inhibition of iRhom1 by CD44-targeting nanocarrier for improved cancer immunochemotherapy.Nat Commun. 2024 Jan 4;15(1):255. doi: 10.1038/s41467-023-44572-6. Nat Commun. 2024. PMID: 38177179 Free PMC article.
-
Applying the logic of genetic interaction to discover small molecules that functionally interact with human disease alleles.Methods Mol Biol. 2015;1263:15-27. doi: 10.1007/978-1-4939-2269-7_2. Methods Mol Biol. 2015. PMID: 25618333 Free PMC article.
References
-
- Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. - PubMed
-
- Arteaga CL. EGF receptor mutations in lung cancer: from humans to mice and maybe back to humans. Cancer Cell. 2006;9:421–423. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300.
-
- Bennett CN, Ross SE, Longo KA, Bajnok L, Hemati N, Johnson KW, Harrison SD, MacDougald OA. Regulation of Wnt signaling during adipogenesis. The Journal of biological chemistry. 2002;277:30998–31004. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

