Centromeric motion facilitates the mobility of interphase genomic regions in fission yeast
- PMID: 23986481
- PMCID: PMC3828595
- DOI: 10.1242/jcs.133678
Centromeric motion facilitates the mobility of interphase genomic regions in fission yeast
Abstract
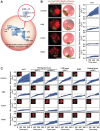
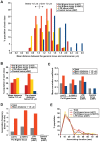
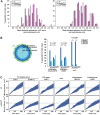
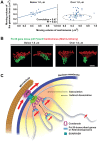
Dispersed genetic elements, such as retrotransposons and Pol-III-transcribed genes, including tRNA and 5S rRNA, cluster and associate with centromeres in fission yeast through the function of condensin. However, the dynamics of these condensin-mediated genomic associations remains unknown. We have examined the 3D motions of genomic loci including the centromere, telomere, rDNA repeat locus, and the loci carrying Pol-III-transcribed genes or long-terminal repeat (LTR) retrotransposons in live cells at as short as 1.5-second intervals. Treatment with carbendazim (CBZ), a microtubule-destabilizing agent, not only prevents centromeric motion, but also reduces the mobility of the other genomic loci during interphase. Further analyses demonstrate that condensin-mediated associations between centromeres and the genomic loci are clonal, infrequent and transient. However, when associated, centromeres and the genomic loci migrate together in a coordinated fashion. In addition, a condensin mutation that disrupts associations between centromeres and the genomic loci results in a concomitant decrease in the mobility of the loci. Our study suggests that highly mobile centromeres pulled by microtubules in cytoplasm serve as 'genome mobility elements' by facilitating physical relocations of associating genomic regions.
Keywords: Condensin; Fission yeast; Genome dynamics; Live-cell imaging; Nuclear organization.
Figures


Similar articles
-
Potential roles of condensin in genome organization and beyond in fission yeast.J Microbiol. 2021 May;59(5):449-459. doi: 10.1007/s12275-021-1039-2. Epub 2021 Apr 20. J Microbiol. 2021. PMID: 33877578 Review.
-
Centromeric localization of dispersed Pol III genes in fission yeast.Mol Biol Cell. 2010 Jan 15;21(2):254-65. doi: 10.1091/mbc.e09-09-0790. Epub 2009 Nov 12. Mol Biol Cell. 2010. PMID: 19910488 Free PMC article.
-
Global genome organization mediated by RNA polymerase III-transcribed genes in fission yeast.Gene. 2012 Feb 10;493(2):195-200. doi: 10.1016/j.gene.2010.12.011. Epub 2010 Dec 30. Gene. 2012. PMID: 21195141 Free PMC article. Review.
-
Interaction between TBP and Condensin Drives the Organization and Faithful Segregation of Mitotic Chromosomes.Mol Cell. 2015 Sep 3;59(5):755-67. doi: 10.1016/j.molcel.2015.07.007. Epub 2015 Aug 6. Mol Cell. 2015. PMID: 26257282 Free PMC article.
-
Fission yeast condensin contributes to interphase chromatin organization and prevents transcription-coupled DNA damage.Genome Biol. 2020 Nov 5;21(1):272. doi: 10.1186/s13059-020-02183-0. Genome Biol. 2020. PMID: 33153481 Free PMC article.
Cited by
-
Nuclear Mechanics in the Fission Yeast.Cells. 2019 Oct 20;8(10):1285. doi: 10.3390/cells8101285. Cells. 2019. PMID: 31635174 Free PMC article. Review.
-
An IF-FISH Approach for Covisualization of Gene Loci and Nuclear Architecture in Fission Yeast.Methods Enzymol. 2016;574:167-180. doi: 10.1016/bs.mie.2016.04.003. Epub 2016 Jun 1. Methods Enzymol. 2016. PMID: 27423862 Free PMC article.
-
Involvement of condensin-directed gene associations in the organization and regulation of chromosome territories during the cell cycle.Nucleic Acids Res. 2016 May 5;44(8):3618-28. doi: 10.1093/nar/gkv1502. Epub 2015 Dec 23. Nucleic Acids Res. 2016. PMID: 26704981 Free PMC article.
-
Potential roles of condensin in genome organization and beyond in fission yeast.J Microbiol. 2021 May;59(5):449-459. doi: 10.1007/s12275-021-1039-2. Epub 2021 Apr 20. J Microbiol. 2021. PMID: 33877578 Review.
-
Transcription factors mediate condensin recruitment and global chromosomal organization in fission yeast.Nat Genet. 2016 Oct;48(10):1242-52. doi: 10.1038/ng.3647. Epub 2016 Aug 22. Nat Genet. 2016. PMID: 27548313 Free PMC article.
References
-
- Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y - DOI - PubMed
-
- Bowen N. J., Jordan I. K., Epstein J. A., Wood V., Levin H. L. (2003). Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements from the complete genome sequence of Schizosaccharomyces pombe. Genome Res. 13, 1984–1997 10.1101/gr.1191603 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

