Metagenomic analysis of the ferret fecal viral flora
- PMID: 23977082
- PMCID: PMC3748082
- DOI: 10.1371/journal.pone.0071595
Metagenomic analysis of the ferret fecal viral flora
Abstract
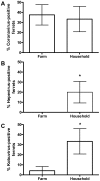
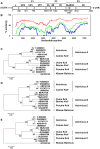
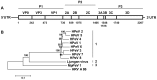
Ferrets are widely used as a small animal model for a number of viral infections, including influenza A virus and SARS coronavirus. To further analyze the microbiological status of ferrets, their fecal viral flora was studied using a metagenomics approach. Novel viruses from the families Picorna-, Papilloma-, and Anelloviridae as well as known viruses from the families Astro-, Corona-, Parvo-, and Hepeviridae were identified in different ferret cohorts. Ferret kobu- and hepatitis E virus were mainly present in human household ferrets, whereas coronaviruses were found both in household as well as farm ferrets. Our studies illuminate the viral diversity found in ferrets and provide tools to prescreen for newly identified viruses that potentially could influence disease outcome of experimental virus infections in ferrets.
Conflict of interest statement
Figures


Similar articles
-
Determination of Ferret Enteric Coronavirus Genome in Laboratory Ferrets.Emerg Infect Dis. 2017 Sep;23(9):1568-1570. doi: 10.3201/eid2309.160215. Emerg Infect Dis. 2017. PMID: 28820366 Free PMC article.
-
Molecular detection and characterization of human gyroviruses identified in the ferret fecal virome.Arch Virol. 2014 Dec;159(12):3401-6. doi: 10.1007/s00705-014-2203-3. Epub 2014 Aug 14. Arch Virol. 2014. PMID: 25119678 Free PMC article.
-
Metagenomic Survey of Viral Diversity Obtained from Feces of Subantarctic and South American Fur Seals.PLoS One. 2016 Mar 17;11(3):e0151921. doi: 10.1371/journal.pone.0151921. eCollection 2016. PLoS One. 2016. PMID: 26986573 Free PMC article.
-
The ferret as a model organism to study influenza A virus infection.Dis Model Mech. 2011 Sep;4(5):575-9. doi: 10.1242/dmm.007823. Epub 2011 Aug 2. Dis Model Mech. 2011. PMID: 21810904 Free PMC article. Review.
-
New strategy for virus discovery: viruses identified in human feces in the last decade.Sci China Life Sci. 2013 Aug;56(8):688-96. doi: 10.1007/s11427-013-4516-y. Epub 2013 Aug 7. Sci China Life Sci. 2013. PMID: 23917840 Free PMC article. Review.
Cited by
-
Duck gut viral metagenome analysis captures snapshot of viral diversity.Gut Pathog. 2016 Jun 9;8:30. doi: 10.1186/s13099-016-0113-5. eCollection 2016. Gut Pathog. 2016. PMID: 27284287 Free PMC article.
-
Structure and function of type IV IRES in picornaviruses: a systematic review.Front Microbiol. 2024 May 24;15:1415698. doi: 10.3389/fmicb.2024.1415698. eCollection 2024. Front Microbiol. 2024. PMID: 38855772 Free PMC article.
-
The Gut Viral Metagenome Analysis of Domestic Dogs Captures Snapshot of Viral Diversity and Potential Risk of Coronavirus.Front Vet Sci. 2021 Jul 7;8:695088. doi: 10.3389/fvets.2021.695088. eCollection 2021. Front Vet Sci. 2021. PMID: 34307533 Free PMC article.
-
A diverse group of small circular ssDNA viral genomes in human and non-human primate stools.Virus Evol. 2015 Dec 31;1(1):vev017. doi: 10.1093/ve/vev017. eCollection 2015. Virus Evol. 2015. PMID: 27774288 Free PMC article.
-
Nasal virome of dogs with respiratory infection signs include novel taupapillomaviruses.Virus Genes. 2019 Apr;55(2):191-197. doi: 10.1007/s11262-019-01634-6. Epub 2019 Jan 10. Virus Genes. 2019. PMID: 30632017 Free PMC article.
References
-
- Kuiken T, Leighton FA, Fouchier RA, LeDuc JW, Peiris JS, et al. (2005) Public health. Pathogen surveillance in animals. Science 309: 1680–1681 309/5741/1680 [pii];10.1126/science.1113310 [doi] - DOI - PubMed
-
- Taylor LH, Latham SM, Woolhouse ME (2001) Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci 356: 983–989 10.1098/rstb.2001.0888 [doi] - DOI - PMC - PubMed
-
- Woolhouse ME, Haydon DT, Antia R (2005) Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol Evol 20: 238–244 S0169–5347(05)00038–8 [pii];10.1016/j.tree.2005.02.009 [doi] - DOI - PMC - PubMed
-
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, et al. (1999) Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397: 436–441 10.1038/17130 [doi] - DOI - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

