ITPA (inosine triphosphate pyrophosphatase): from surveillance of nucleotide pools to human disease and pharmacogenetics
- PMID: 23969025
- PMCID: PMC3827912
- DOI: 10.1016/j.mrrev.2013.08.001
ITPA (inosine triphosphate pyrophosphatase): from surveillance of nucleotide pools to human disease and pharmacogenetics
Abstract
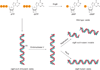
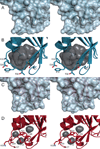
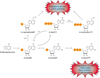
Cellular nucleotide pools are often contaminated by base analog nucleotides which interfere with a plethora of biological reactions, from DNA and RNA synthesis to cellular signaling. An evolutionarily conserved inosine triphosphate pyrophosphatase (ITPA) removes the non-canonical purine (d)NTPs inosine triphosphate and xanthosine triphosphate by hydrolyzing them into their monophosphate form and pyrophosphate. Mutations in the ITPA orthologs in model organisms lead to genetic instability and, in mice, to severe developmental anomalies. In humans there is genetic polymorphism in ITPA. One allele leads to a proline to threonine substitution at amino acid 32 and causes varying degrees of ITPA deficiency in tissues and plays a role in patients' response to drugs. Structural analysis of this mutant protein reveals that the protein is destabilized by the formation of a cavity in its hydrophobic core. The Pro32Thr allele is thought to cause the observed dominant negative effect because the resulting active enzyme monomer targets both homo- and heterodimers to degradation.
Keywords: Base analogs; DSB; Dominant negative; HAM1; HAP; HGPRT; ITP; ITPA; ITPA gene polymorphism; MEF; Mercaptopurines; NUDT16; Nucleotide pool; Pharmacogenetics; Protein stability; SSB; Saccharomyces cerevisiae; TPMT; XTP; base analog 6-hydroxylaminopurine; double-strand break; homolog RdgB E. coli ITPA homolog (Rec-dependent growth B); hypoxanthine-guanine phosphoribosyltransferase; inosine triphosphate; inosine triphosphate pyrophosphatase; mouse embryonic fibroblasts; nudix (nucleoside diphosphate linked moiety X)-type motif 16; single-strand break; thiopurine S-methyltransferase; xanthosine triphosphate.
Copyright © 2013 Elsevier B.V. All rights reserved.
Conflict of interest statement
None of the contributors to this article has any conflicting interests.
Figures
Similar articles
-
The human ITPA polymorphic variant P32T is destabilized by the unpacking of the hydrophobic core.J Struct Biol. 2013 Jun;182(3):197-208. doi: 10.1016/j.jsb.2013.03.007. Epub 2013 Mar 23. J Struct Biol. 2013. PMID: 23528839 Free PMC article.
-
Functional study of the P32T ITPA variant associated with drug sensitivity in humans.J Mol Biol. 2009 Sep 25;392(3):602-13. doi: 10.1016/j.jmb.2009.07.051. Epub 2009 Jul 23. J Mol Biol. 2009. PMID: 19631656 Free PMC article.
-
Inosine Triphosphate Pyrophosphatase (ITPase): Functions, Mutations, Polymorphisms and Its Impact on Cancer Therapies.Cells. 2022 Jan 24;11(3):384. doi: 10.3390/cells11030384. Cells. 2022. PMID: 35159194 Free PMC article. Review.
-
Pivotal role of inosine triphosphate pyrophosphatase in maintaining genome stability and the prevention of apoptosis in human cells.PLoS One. 2012;7(2):e32313. doi: 10.1371/journal.pone.0032313. Epub 2012 Feb 27. PLoS One. 2012. PMID: 22384212 Free PMC article.
-
Inosine triphosphate pyrophosphatase: A guardian of the cellular nucleotide pool and potential mediator of RNA function.Wiley Interdiscip Rev RNA. 2023 Sep-Oct;14(5):e1790. doi: 10.1002/wrna.1790. Epub 2023 Apr 24. Wiley Interdiscip Rev RNA. 2023. PMID: 37092460 Review.
Cited by
-
ITPA, TPMT, and NUDT15 Genetic Polymorphisms Predict 6-Mercaptopurine Toxicity in Middle Eastern Children With Acute Lymphoblastic Leukemia.Front Pharmacol. 2019 Aug 27;10:916. doi: 10.3389/fphar.2019.00916. eCollection 2019. Front Pharmacol. 2019. PMID: 31507415 Free PMC article.
-
Clinico-radiological features, molecular spectrum, and identification of prognostic factors in developmental and epileptic encephalopathy due to inosine triphosphate pyrophosphatase (ITPase) deficiency.Hum Mutat. 2022 Mar;43(3):403-419. doi: 10.1002/humu.24326. Epub 2022 Jan 12. Hum Mutat. 2022. PMID: 34989426 Free PMC article.
-
Genomic analysis of a riboflavin-overproducing Ashbya gossypii mutant isolated by disparity mutagenesis.BMC Genomics. 2020 Apr 23;21(1):319. doi: 10.1186/s12864-020-6709-7. BMC Genomics. 2020. PMID: 32326906 Free PMC article.
-
A Necroptosis-Related Prognostic Model of Uveal Melanoma Was Constructed by Single-Cell Sequencing Analysis and Weighted Co-Expression Network Analysis Based on Public Databases.Front Immunol. 2022 Feb 15;13:847624. doi: 10.3389/fimmu.2022.847624. eCollection 2022. Front Immunol. 2022. PMID: 35242144 Free PMC article.
-
Inosine triphosphate pyrophosphatase from Trypanosoma brucei cleanses cytosolic pools from deaminated nucleotides.Sci Rep. 2022 Apr 18;12(1):6408. doi: 10.1038/s41598-022-10149-4. Sci Rep. 2022. PMID: 35436992 Free PMC article.
References
-
- Lin S, McLennan AG, Ying K, Wang Z, Gu S, Jin H, Wu C, Liu W, Yuan Y, Tang R, Xie Y, Mao Y. Cloning, expression, and characterization of a human inosine triphosphate pyrophosphatase encoded by the ITPA gene. J. Biol. Chem. 2001;276:18695–18701. - PubMed
-
- Pavlov YI. Saccharomyces cerevisiae mutants highly sensitive to the mutagenic action of 6-N-hydroxylaminopurine. Sov. Genet. 1986;22:1099–1107.
-
- Behmanesh M, Sakumi K, Abolhassani N, Toyokuni S, Oka S, Ohnishi YN, Tsuchimoto D, Nakabeppu Y. ITPase-deficient mice show growth retardation and die before weaning. Cell Death Differ. 2009;16:1315–1322. - PubMed
-
- Marinaki AM, Ansari A, Duley JA, Arenas M, Sumi S, Lewis CM, Shobowale-Bakre e-M, Escuredo E, Fairbanks LD, Sanderson JD. Adverse drug reactions to azathioprine therapy are associated with polymorphism in the gene encoding inosine triphosphate pyrophosphatase (ITPase) Pharmacogenetics. 2004;14:181–187. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

