Chemical tools for temporally and spatially resolved mass spectrometry-based proteomics
- PMID: 23943069
- PMCID: PMC3925203
- DOI: 10.1007/s10439-013-0878-3
Chemical tools for temporally and spatially resolved mass spectrometry-based proteomics
Abstract
Accurate measurements of the abundances, synthesis rates and degradation rates of cellular proteins are critical for understanding how cells and organisms respond to changes in their environments. Over the past two decades, there has been increasing interest in the use of mass spectrometry for proteomic analysis. In many systems, however, protein diversity as well as cell and tissue heterogeneity limit the usefulness of mass spectrometry-based proteomics. As a result, researchers have had difficulty in systematically identifying proteins expressed within specified time intervals, or low abundance proteins expressed in specific tissues or in a few cells in complex microbial systems. In this review, we present recently-developed tools and strategies that probe these two subsets of the proteome: proteins synthesized during well-defined time intervals--temporally resolved proteomics--and proteins expressed in predetermined cell types, cells or cellular compartments--spatially resolved proteomics--with a focus on chemical and biological mass spectrometry-based methodologies.
Figures
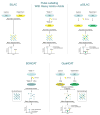



Similar articles
-
Recent trends of capillary electrophoresis-mass spectrometry in proteomics research.Mass Spectrom Rev. 2019 Nov;38(6):445-460. doi: 10.1002/mas.21599. Epub 2019 Aug 12. Mass Spectrom Rev. 2019. PMID: 31407381 Free PMC article. Review.
-
Mass spectral analysis in proteomics.Annu Rev Biophys Biomol Struct. 2004;33:297-316. doi: 10.1146/annurev.biophys.33.111502.082538. Annu Rev Biophys Biomol Struct. 2004. PMID: 15139815 Review.
-
Recent progress in mass spectrometry proteomics for biomedical research.Sci China Life Sci. 2017 Oct;60(10):1093-1113. doi: 10.1007/s11427-017-9175-2. Epub 2017 Oct 13. Sci China Life Sci. 2017. PMID: 29039124 Review.
-
Advances in shotgun proteomics and the analysis of membrane proteomes.J Proteomics. 2010 Oct 10;73(11):2078-91. doi: 10.1016/j.jprot.2010.08.005. Epub 2010 Aug 23. J Proteomics. 2010. PMID: 20797458 Review.
-
Developments in quantitative mass spectrometry for the analysis of proteome dynamics.Trends Biotechnol. 2012 Dec;30(12):668-76. doi: 10.1016/j.tibtech.2012.09.007. Epub 2012 Oct 27. Trends Biotechnol. 2012. PMID: 23107010 Review.
Cited by
-
Tagging and Enriching Proteins Enables Cell-Specific Proteomics.Cell Chem Biol. 2016 Jul 21;23(7):805-815. doi: 10.1016/j.chembiol.2016.05.018. Cell Chem Biol. 2016. PMID: 27447048 Free PMC article.
-
Combining random gene fission and rational gene fusion to discover near-infrared fluorescent protein fragments that report on protein-protein interactions.ACS Synth Biol. 2015 May 15;4(5):615-24. doi: 10.1021/sb5002938. Epub 2014 Oct 14. ACS Synth Biol. 2015. PMID: 25265085 Free PMC article.
-
MACSPI enables tissue-selective proteomic and interactomic analyses in multicellular organisms.Proc Natl Acad Sci U S A. 2024 May 21;121(21):e2319060121. doi: 10.1073/pnas.2319060121. Epub 2024 May 16. Proc Natl Acad Sci U S A. 2024. PMID: 38753516 Free PMC article.
-
Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids.Nat Biotechnol. 2015 Dec;33(12):1272-1279. doi: 10.1038/nbt.3372. Epub 2015 Nov 16. Nat Biotechnol. 2015. PMID: 26571098 Free PMC article.
-
Fluorescent imaging of protein myristoylation during cellular differentiation and development.J Lipid Res. 2017 Oct;58(10):2061-2070. doi: 10.1194/jlr.D074070. Epub 2017 Jul 28. J Lipid Res. 2017. PMID: 28754825 Free PMC article.
References
-
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. - PubMed
-
- Agard N, Prescher J, Bertozzi C. A strain-promoted [3+2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc. 2004;126:15046–15047. - PubMed
-
- Ahrens CH, Brunner E, Qeli E, Basler K, Aebersold R. Generating and navigating proteome maps using mass spectrometry. Nat Rev Mol Cell Biol. 2010;11:789–801. - PubMed
-
- Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389:1017–1031. - PubMed
-
- Bantscheff M, Lemeer S, Savitski MH, Kuster B. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal Bioanal Chem. 2012;404:939–965. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

